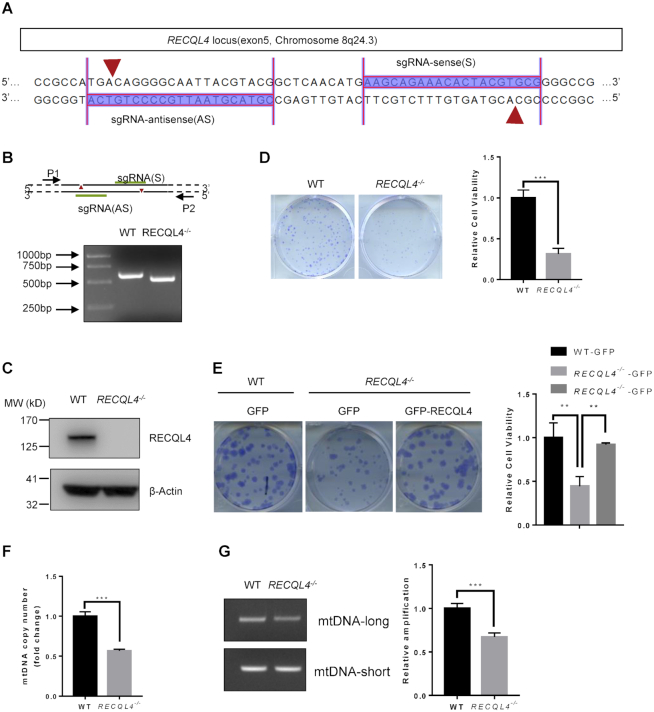
Figure 1.
Characterization of RECQL4 knockout cells. (A) Schematic showing sgRNA sequences targeting the human RECQL4 locus using Cas9 D10A nickases (Cas9n). Red arrows indicate the sites of nicks in DNA. (B) PCR analyses of WT and RECQL4−/− cells using primer pair (P1+P2) to amplify the genomic region ranging from exon 5 to intron 6 of RECQL4. The result shows the RECQL4−/− clone with a homozygous deletion of the RECQL4 locus. (C) Knockout of RECQL4 in U2OS cells was confirmed by immunoblotting. (D) Colony formation assay showing RECQL4−/− cells have clonogenic survival defects. Images show representative colonies stained with crystal violet. Data are shown as mean ± SEM, n = 3. ***P < 0.001 (t-test). (E) Colony formation assay showing that re-introduction of WT-RECQL4 in RECQL4−/− cells rescues the clonogenic survival defects of RECQL4−/-cells. Three independent experiments were performed, each with four replicates. Representative images (left) and quantifications (right) are shown. **P < 0.01, using unpaired two-tailed Student's t test. (F) mtDNA copy number in WT and RECQL4−/− cells. Data are shown as mean ± SEM, n = 3. ***P < 0.001 (t-test). (G) mtDNA damage detection by a PCR-based assay assessing relative quantitative amplification of a small (117 bp) to a large (10 kb) segment of mtDNA. A representative gel (left) and quantifications (right) are shown as mean ± SEM, n = 3. ***P < 0.001.