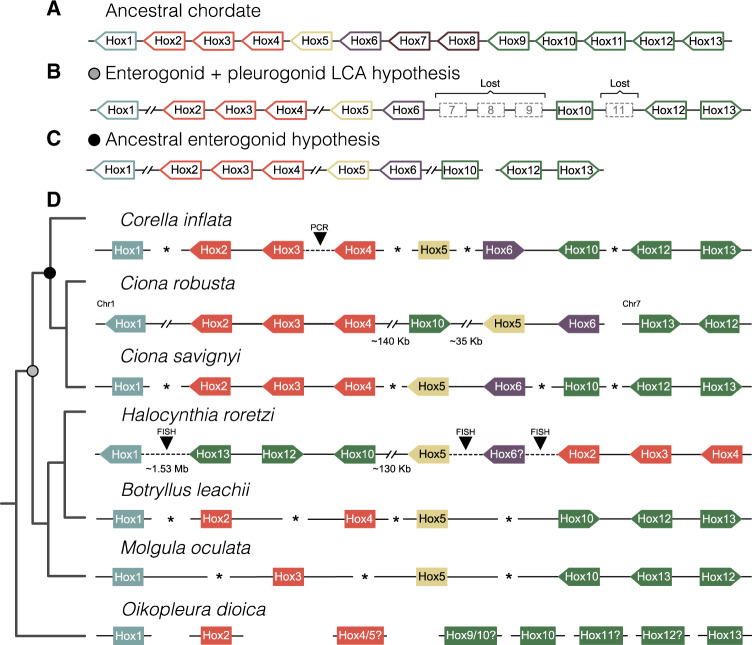
Fig. 5.
—Genomic organization of Hox genes in tunicates and the chordate ancestor. Linked Hox genes are connected by solid lines. Dashed lines indicate Hox genes that are currently located on separate genomic scaffolds but were shown to be linked using other methods (e.g., FISH, PCR). Asterisks between Hox genes indicate that linkage is unknown. The distances between Hox genes are not to scale. Distances of at least 35 kb are indicated with paired forward slashes. If known, the transcription direction for linked genes is indicated by the direction of the arrow. Non-Hox genes that may be present between Hox genes are not shown. Chromosome numbers and linkage information for Ciona robusta are from Satou et al. (2019). (A) Hox cluster in the ancestral chordate. (B) Inferred Hox cluster in the last common ancestor of enterogonid and enterogonid tunicates. The gray circle represents the position of this ancestral in the tunicate tree. (C) Inferred Hox cluster in the enterogonid ancestor. The black circle represents the position of the ancestral enterogonid in the tunicate tree. (D) Linkage information for extant tunicates. The linkage shown here for Ci. robusta is notably different from that in Blanchoud et al. (2018) who did not report the FISH results from Ikuta et al. (2004). The cladogram on the left shows the evolutionary relationships between taxa. Scaffold identification numbers and sequence coordinates for tunicate Hox genes are available in supplementary table S3, Supplementary Material online.