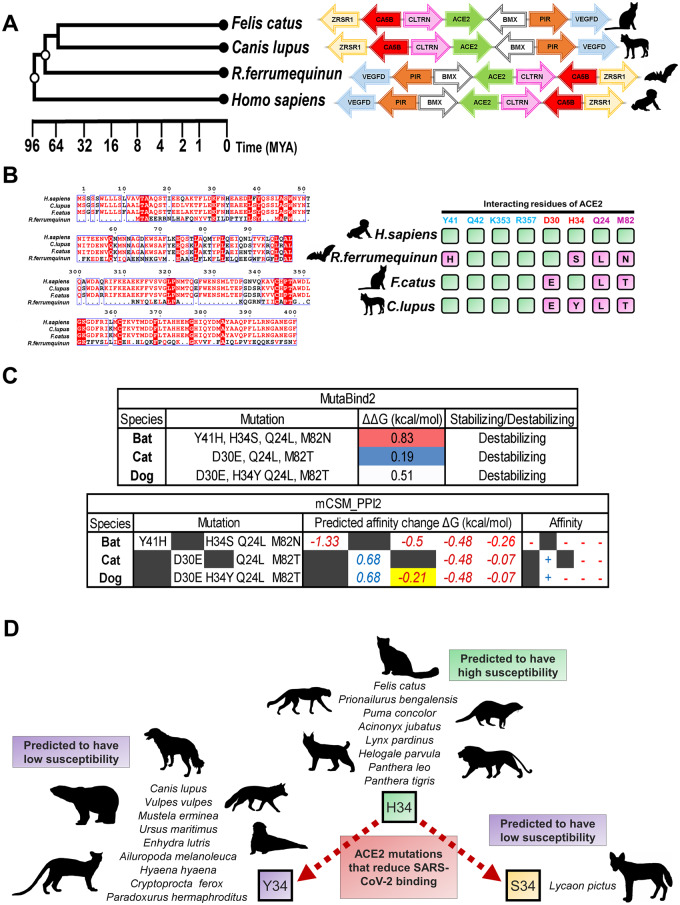
Figure 1.
(A) The gene locus of ACE2 is highly conserved across vertebrates. Phylogeny of cats (F.catus), dogs (C.lupus), greater horseshoe bat (Rhinolophus ferrumequinum) and humans. Tree and estimated times of divergence (MYA, millions of years ago) were obtained from TimeTree (http://www.timetree.org). For each described species, the ACE2 ortholog gene loci are indicated and conserved genes within the syntenic region are color coded. UCSC Genome Browser (https://genome.ucsc.edu) was used to view the respective of ACE2 loci of each species. Arrows indicate the direction of transcription predicted by UCSC Genome Browser annotations. (B) Alignment of ACE2 protein and its orthologs reveals differences in key residues targeted by SARS-CoV-2. Protein sequences of F.catus, C.lupus, R.ferrumequinum ACE2 were aligned using MUSCLE alignment on the MEGA7 tool (https://www.megasoftware.net/) and sequences from positions 0 to 100 and 300 to 400 are shown. Display for the alignment was generated using the ESPRIPT 3.0 tool (http://espript.ibcp.fr/). Interaction residues between ACE2 and the SARS-CoV-2 spike protein (PDB: 6M17) identified by Yan et al.[2] are mapped to the ACE2 orthologs and compared to the human ACE2 green=conserved, magenta=mutated and the letter indicates the residue present in the ortholog. (C) H34Y is predicted to reduce the binding affinity between ACE2 and SARS-CoV-2. MutaBind2 (https://lilab.jysw.suda.edu.cn/research/mutabind2/) and mCSM-PPI2 (http://biosig.unimelb.edu.au/mcsm_ppi2/) analyses reveal changes from mutations in binding affinity between the ACE2 receptor and RBD of the SARS-CoV-2 spike protein. Analyses described in the main text of the article. Affinity indicates the change associated with each binding energy ‘+’ being stabilizing and ‘−’ being destabilizing. (D) H34 mutations in Carnivora predict the susceptibility of species. We analyzed ACE2 orthologs based on sequences from available genomes from Feliforma and Caniforma suborders to determine which species have H34Y mutations. These species with H34S and H34Y mutations are predicted to have reduced susceptibility to the SARS-CoV-2 virus. Silhouettes of organisms from each representative species were obtained from PhyloPic (http://phylopic.org)