Figure 3.
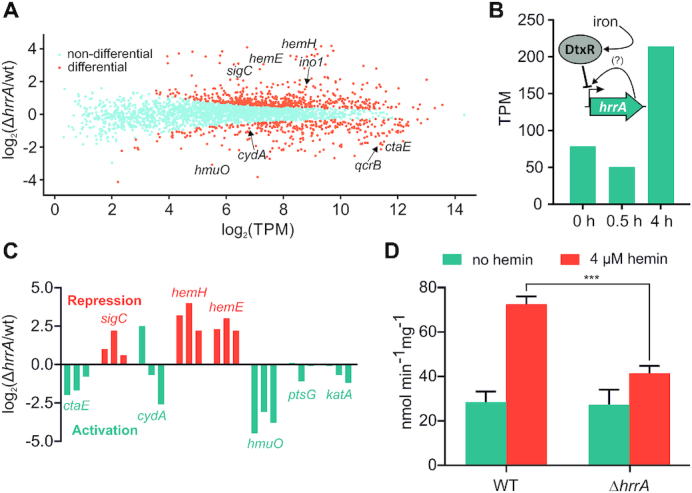
Differential gene expression analysis of wild-type Corynebacterium glutamicum and a ΔhrrA mutant. (A) Differential gene expression analysis (RNA-Seq) revealed 120 upregulated and 154 downregulated genes in the hrrA deletion strain compared to the wild-type (in transcripts per million, TPM) after 30 min of cultivation in iron-depleted glucose minimal medium containing 4 μM heme. (B) Expression levels of hrrA (TPM) 0, 0.5 and 4 h after the addition of hemin. A scheme depicts HrrA autoregulation and iron-dependent DtxR repression (24). (C) Impact of hrrA deletion on the transcript levels of six selected target genes at three different time points (0, 0.5, 4 h; orange: HrrA acts as a repressor, turquoise: HrrA acts as an activator). (D) Measurement of cytochrome aa3 oxidase activity using the TMPD oxidase assay in C. glutamicum wild-type and ΔhrrA grown with or without 4 μM heme.
