Figure 2.
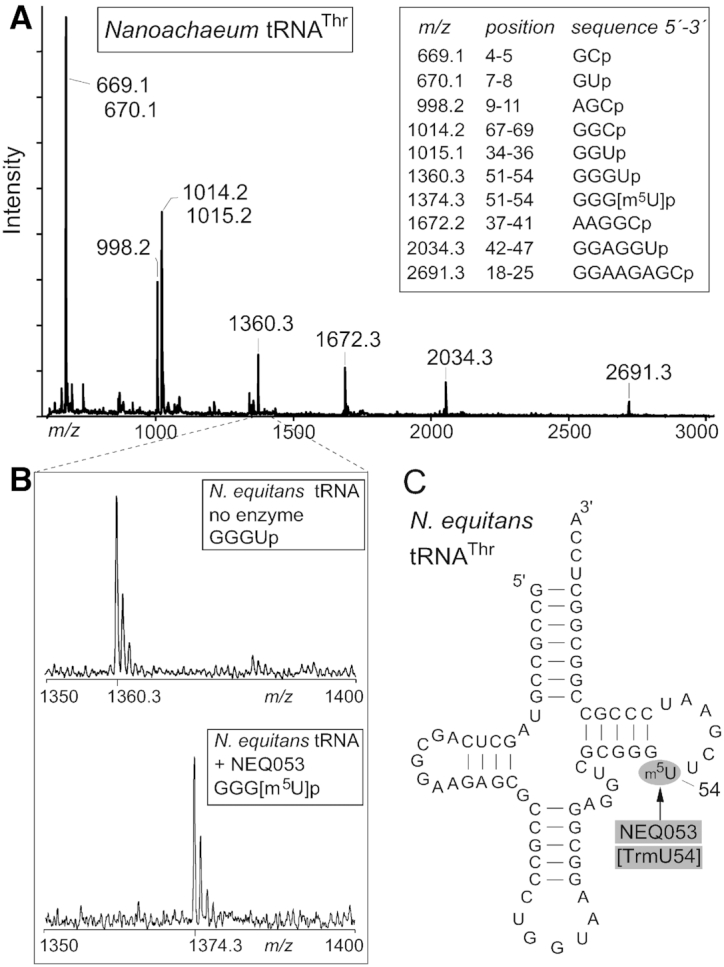
MALDI-MS spectra showing methylation in vitro by NEQ053 at U54 in N. equitans tRNAThr. (A) Spectrum of oligonucleotides formed by RNase A digestion of unmodified tRNAThr. The measured mass/charge (m/z) of each fragment is shown above the peak with the theoretical m/z values (in box). (B) Enlargement of the spectral region containing the GGGUp fragment from nucleotides 51–54 with an m/z of 1360.3 when derived from the unmodified tRNAThr, and with m/z of 1374.3 after incubation of the tRNA in vitro with NEQ053 prior to digestion. (C) Schematic of the N. equitans tRNAThr secondary structure. The target site of NEQ053 indicates that the enzyme could be renamed as TrmU54.
