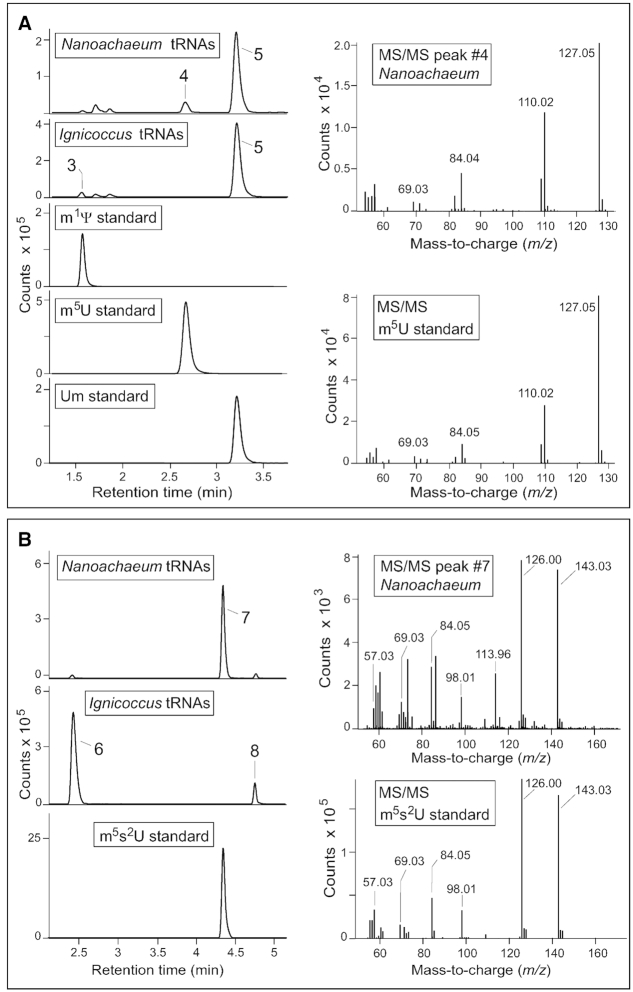
Figure 3.
LC/ESI-MS characterization of uridine nucleosides from N. equitans and I. hospitalis tRNAs. (A) Analyses of uridines modified with one methyl group and registered at m/z 259.09 in positive ion mode. On left, retention times during liquid chromatography compared to known methyluridine standards. On right, fragmentation of N. equitans compound #4 confirming its structure from comparison with the m5U standard. The identities of compound #3 (m1Ψ) and compound #5 (Um) were verified by the same methods (Supplementary Figure S3). Minor amounts of non-nucleoside compounds can be seen migrating at retention times close to that of m1Ψ. (B) Uridines modified with a methyl plus a thiol group registered at m/z 275.07 in positive ion mode. Compound #7 is specific to N. equitans and matches the LC profile of the m5s2U standard as well as its MS/MS fragmentation pattern (right). No standards with the same LC migration were available for compounds #6 and #8, and these were identified respectively as m1s4Ψ and s2Um by comparing their fragmentation patterns to a series of nucleosides containing these individual modifications (Figures 4 and 5). Small quantities of these compounds were observed in some preparations of the enriched N. equitans cells (as seen here) and are contaminants resulting from incomplete removal of all the I. hospitalis cells.