Figure 6.
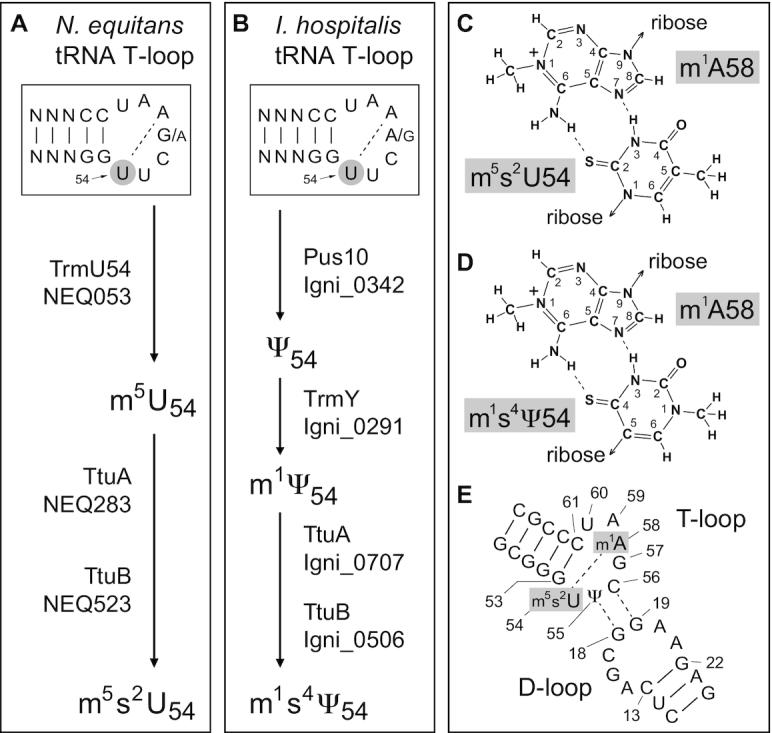
Modification pathways of nucleotide U54 within the tRNAs of (A) N. equitans and (B) I. hospitalis. Modified U54 makes a reverse Hoogsteen interaction with nucleotide 58 in the T-loops of both archaea where (C) the m5s2U54 to m1A58 interaction in N. equitans is functionally equivalent to (D) the m1s4Ψ54 to m1A58 pair found in I. hospitalis. (E) Putative interaction of the archaeal tRNA D- and T-loops based on the yeast tRNAPhe structure (33). The N. equitans and I. hospitalis genomes respectively encode the m1A methyltransferase TrmI homologs Igni_1131 and NEQ337. The presence of m1A was confirmed by LC (retention time of 3.6 min) and by ESI-MS/MS (not shown) in the tRNAs of both archaea; m1I (LC retention time of 3.6 min) was also found in both species, and presumably is formed at position 57 via an m1A intermediate in tRNAs that have an adenosine at this position (67,68). MS fragmentation confirmed the structures of Ψ (Figure 5), m5s2U54 (Figure 3) and m1s4Ψ54 (Figures 4 and 5).
