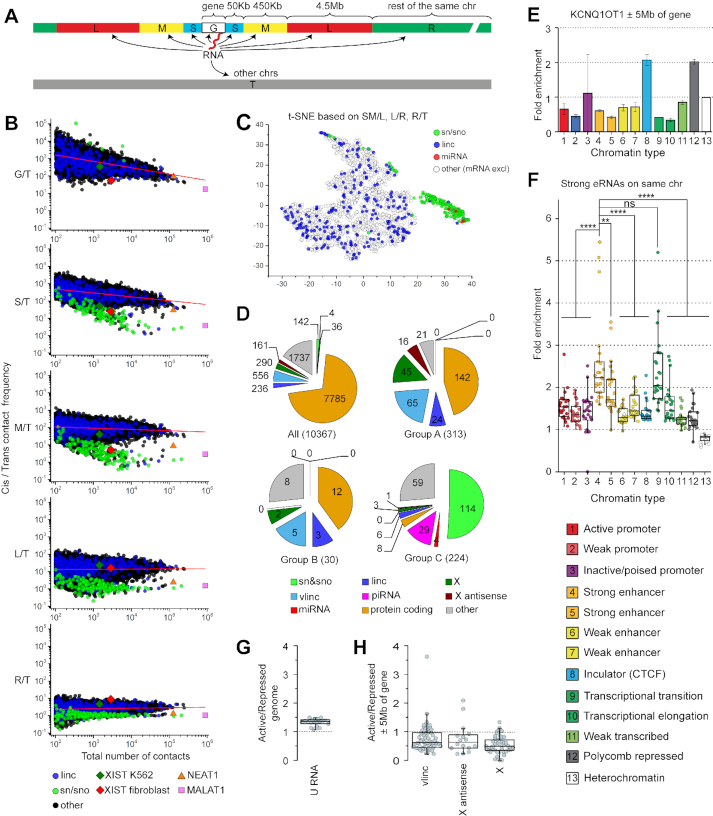
Figure 2.
Preferences of RNAs for short- and long-range contacts and different chromatin types in K562 cells. (A) Scheme demonstrating analyzed genomic intervals. (B) Ratio of contact frequency of individual RNAs with regions of parental chromosome to contact frequency of the same RNAs with the other chromosomes (Y axis) versus total number of contacts (X axis). Graphs from top to bottom show results for different cis intervals as specified in (A). RNAs with ≥ 100 contacts are presented. Red line, linear regression. (C) T-SNE analysis of RNAs based on ratios between contact frequencies in consecutive intervals. (D) Number of RNAs of a particular biotype in group A (RNAs enriched in gene-proximal areas), group B (XIST-like RNAs), group C (RNAs distributed throughout the genome), and among all analyzed RNAs. (E) Fold enrichment of Kcnq1ot1 at specific chromatin types in the region surrounding Kcnq1ot1 gene (± 5 Mb of gene boundaries) relative to overall contact frequency in this region. Error bars, SEM for two biological replicates. (F) Fold enrichment of eRNAs produced from chromatin type 4 and 5 at specific chromatin types within the same chromosome relative to overall contact frequency in the same chromosome. Points represent results for individual chromosomes (n = 23, P-values are from Tukey's multiple comparisons test). (G, H) Ratio between contact frequencies in active and repressed chromatin for U RNAs belonging to group C (G) and for vlinc, X RNAs, and antisense X RNAs belonging to group A (H). Active chromatin is defined as combination of types 1, 2, 4, 5, 6, 7, 9, 10 and 11; repressed chromatin, of types 3, 12 and 13. Contact frequency was determined for the full genome (G) or in regions ±5Mb of gene boundaries (H).