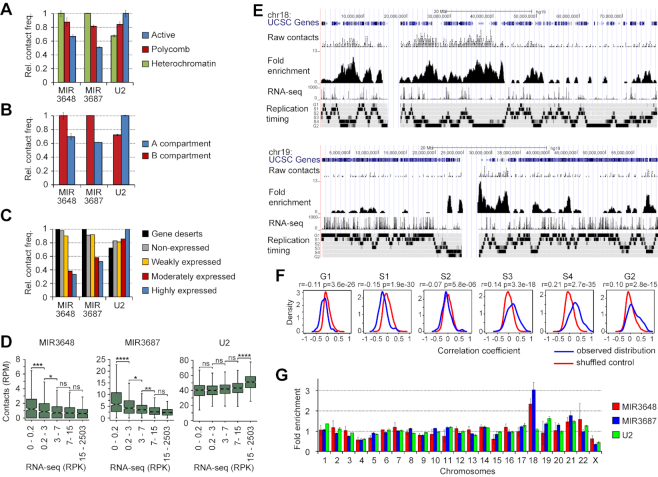
Figure 3.
MIR3648 and MIR3687 target inactive chromatin. (A, B) Frequency of contacts of MIR3648, MIR3687, and U2 with different chromatin types (A) and A and B spatial compartments (B) determined for the full genome. The maximal contact frequency for a given RNA is taken to be equal to 1. Error bars, SEM for two biological replicates. Active chromatin is defined as combination of types 1, 2, 4, 5, 6, 7, 9, 10 and 11; Polycomb, of types 3 and 12; Heterochromatin, of type 13. A/B compartment track for K562 was obtained from (33). (C) Frequency of contacts of MIR3648, MIR3687 and U2 with expressed protein-coding genes (divided into three equal groups based on the density of RNA-seq signal), non-expressed protein-coding genes (RNA-seq signal = 0), and gene deserts (regions of >500 kb not occupied by any genes). For each RNA, the total number of contacts with genes of each group and gene deserts was determined, normalized by the total length of genes in the group and gene deserts, and presented relative to the maximal value for a given RNA (taken equal to 1). (D) Contacts of MIR3648, MIR3687, and U2 with 1 Mb genomic bins divided into five equal groups based on RNA-seq signal in the bin (n = 576, P-values are from Tukey's multiple comparisons test). Bins occupied by chromatin types 1–13 by less than 10% are not included in the analysis. RPK, reads per Kb; RPM, reads per Mb. (E) Distribution of raw contacts of MIR3687 along Chrs 18 and 19 and fold enrichment compared to background at a 50 kb resolution. Gene distribution, RNA-seq signal (1 kb bin), and replication timing profile for K562 as determined by Repli-seq (56) are shown. (F) Distribution of correlation coefficients upon comparison of MIR3687 fold enrichment profile with Repli-seq in genomic windows of 20 Mb, as examined by StereoGene (57). The genome-wide correlation coefficients calculated with the kernel and P-values are presented. (G) Fold enrichment of MIR3648, MIR3687 and U2 at individual chromosomes relative to overall contact frequency of respective RNAs in the genome. Error bars, SEM for two biological replicates.