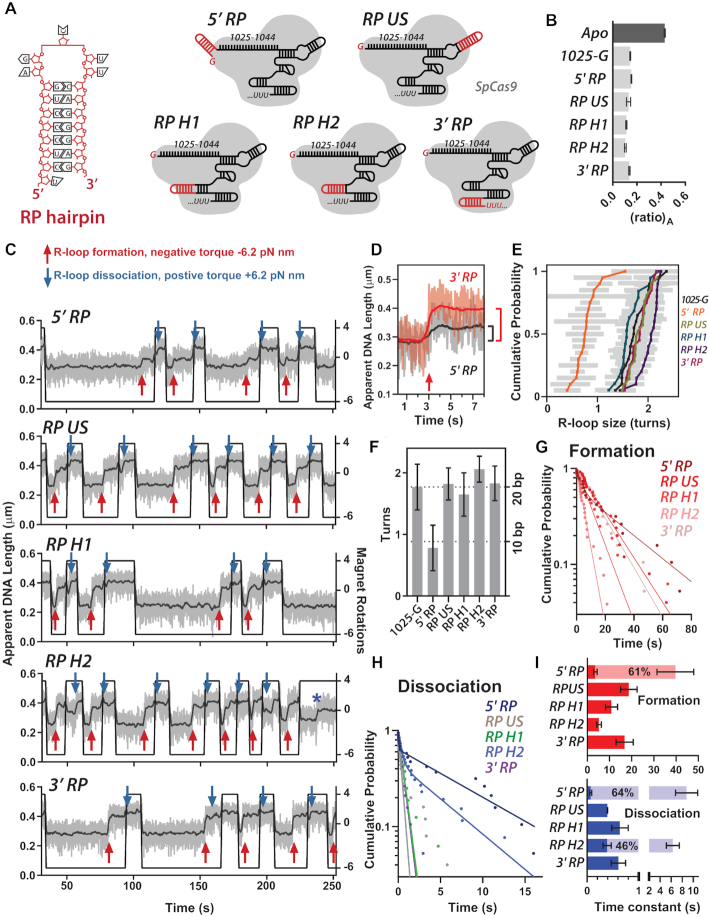
Figure 5.
The effect of the RP RNA hairpin on R-loop dynamics. (A) Sequence of the RP hairpin and schematics of Streptococcus pyogenes Cas9 (grey) and the in vitro transcribed gRNAs (black) (sequences in Supplementary Figure S1). (B) FRET-based gRNA loading assay. Comparative (ratio)A for loading gRNAs as shown (light grey) relative to Apo Cas9 (dark grey) (N = 3 ± SD). (C) Example MT traces of R-loop cycling (at 10 turns s−1) to measure R-loop formation (red arrows) and dissociation (blue arrows). Raw (grey) and 2 Hz smoothed data (black) are shown. (D) Comparison of an R-loop formation event with 3′ RP (red) and 3′ RP (black) gRNA. (E) cumulative probability of R-loop size in turns estimated from R-loop cycling experiments in Supplementary Figure S8 (Materials and Methods). Grey bars are the standard error for each estimation. (F) Average R-loop size in turns (±SD) for each of the gRNAs. The average turns for 1025-G, RP US, RP H1, RP H2 and 3′ RP was assumed to estimate a full length R-loop and used to set R-loop size in bp. Significance test results in Supplementary Table S5. (G, H) Inverted cumulative probability distributions for the R-loop formation and dissociation times for each gRNA. Solid lines are single exponential fits or double exponential fits (5′ RP formation; 5′ RP and RP H2 dissociation). (I) Mean R-loop formation and dissociation times and standard error (N = 19 to 43, Supplementary Table S3) from the fits in panel D. For 5′ RP formation events, and 5′ RP and RP H2 dissociation events, the light and dark bars are the two constants from a double exponential fit with the percentage shown for the amplitude of the slower events. For the dissociation times, the x-axis is split as 0–1 s and 2–10 s.