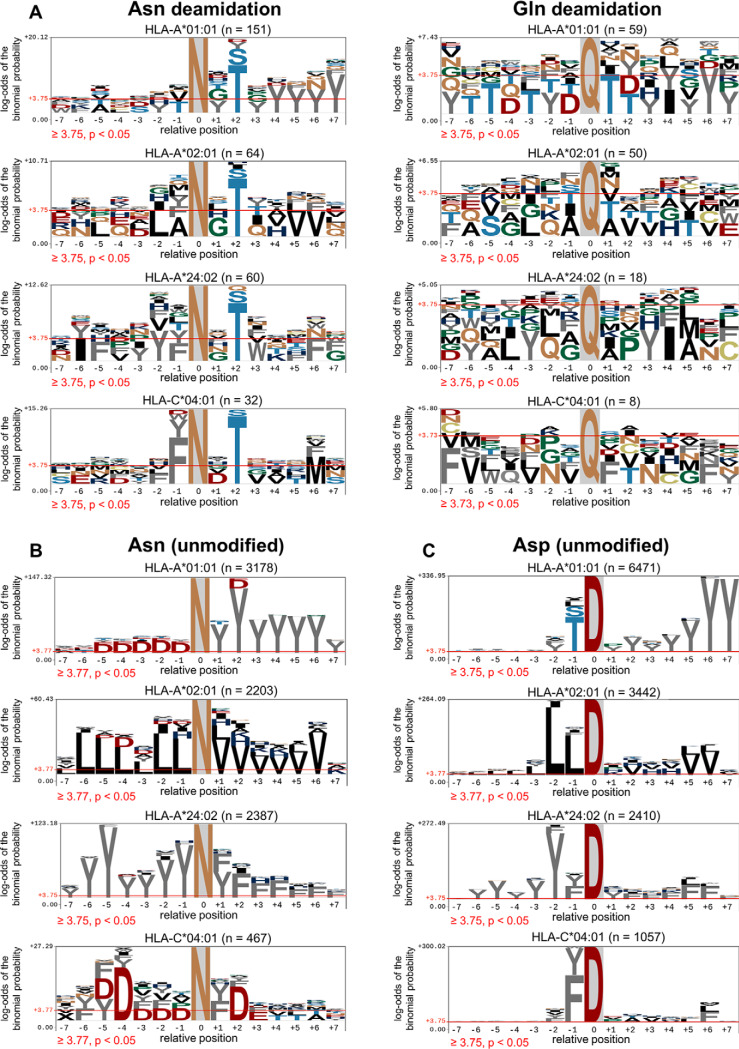
Fig. 3.
Enrichment of amino acid residues flanking the NX(S/T) motif in Asparagine deamidated peptides. A, Flanking residue analysis on asparagine or glutamine deamidated peptides. B, Flanking residue motif analysis of peptides containing unmodified asparagine. C, Flanking residue analysis of peptides containing native aspartic acid. The motif analysis was generated using pLogo (81). Amino acids above the red threshold line indicate the log-odds of the binomial probability is significant (p < 0.05).