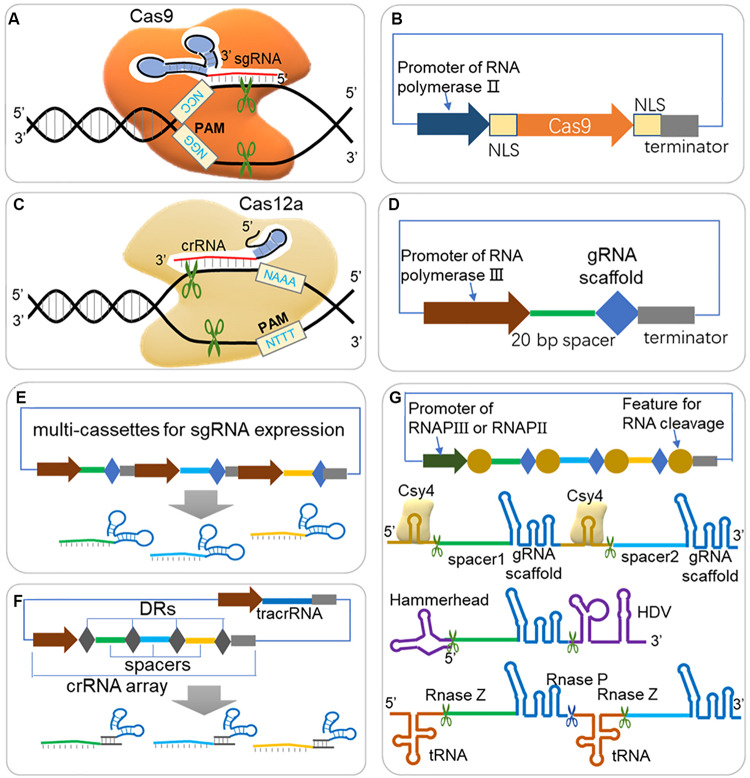
FIGURE 1.
Guidelines for expression of Cas protein and sgRNA in CRISPR/Cas system. (A) Scheme of CRISPR/Cas9 system. The Cas9-sgRNA (or Cas9-crRNA-tracrRNA) complex binds to DNA target arising from Watson-Crick base pairing of spacer sequence, and triggers double strand break (DSB) when next to a short protospacer adjacent motif (PAM, ‘NGG’ for Cas9 from S. pyogenes). (B) Expression cassette for Cas9. For efficient targeting to nucleus in eukaryotes, the Cas9 should be fused to NLS (nuclear localization sequence) at one end or both ends. (C) Scheme of CRISPR/Cas12a (Cpf1) system. Cas12a triggers DSB through a similar scheme of Cas9, but depends on different PAM (‘NTTT’) and less folded crRNA, and creates a sticky end at 18–23 bases away from the PAM. (D) Expression cassette for sgRNA. A promoter of RNA polymerase III (RNAP III) is usually required for directing sgRNA in nucleus and with less modification. A 20 bp spacer should be well designed according to target DNA sequence for efficient editing rates and avoiding off-target effects. (E) Multi-sgRNA expression through multi-cassettes. Repeated elements, such as promoters, gRNA scaffold and terminators are repeated for different spacer sequences. (F) Multi-sgRNA expression through crRNA array and tracrRNA (HI-CRISPR system). Different spacers are separated with direct repeats (DRs) and expressed by one promoter of RNAP III. The pre-crRNA is transcribed and processed into mature crRNA by RNase III and unknown nuclease(s). The tracrRNA and Cas9 protein are complexed with mature crRNA to form the dual-RNA-guided nuclease. (G) gRNA multiplexing strategies. Both RNAP II and RNAP III promoter can be used for expression the sgRNA array, where sgRNAs are separated by features for RNA cleavage. RNA endonuclease Csy4 recognizes a 28 nucleotide sequence flanking the sgRNA sequence and cleaves after the 20th nucleotide. The hammerhead ribozyme and HDV ribozyme flanked the 5′ and 3′ of the sgRNA, respectively, allowing for self-cleaving production of sgRNAs, which are not dependent on the presence of an exogenous protein. Polycistronic tRNA-gRNA architecture allows the production of multiple sgRNAs by endogenous RNase P and RNase Z.