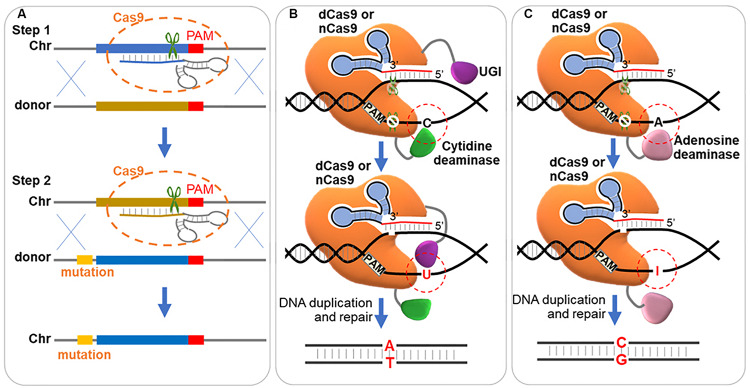
FIGURE 5.
Strategies for precise single base editing. (A) Two-step stuffer-assisted point mutation. In the first step, a 20-nucleotide target genome sequence close to the target is replaced by a heterologous stuffer fragment via homologous recombination. In the second step, the stuffer fragment acts as the target sequence, recognized by a second gRNA, and the original sequence with arbitrary mutation is inserted back. Chr, chromosome. (B) ‘C→T’ mutation through DSB independent pathway. The dCas9 or Cas9 nickase (nCas9, D10A) is fused with cytidine deaminase and uracil DNA glycosylase inhibitor (UGI), and binds to a DNA target. Cytidine deaminase converts the cytidine (‘C’) to uracil (‘U’) in the non-targeted strand, which is protected by UGI from the nucleotide excision repair (NER) pathway. And in the next replication cycle, the ‘G:C’ base pair is repaired to ‘T:A’. (C) ‘A→G’ mutation through DSB independent pathway. The dCas9 or nCas9 is fused with adenosine deaminase and binds to a DNA target. Adenosine deaminase converts the adenosine (‘A’) to hypoxanthine (‘I’) in the non-targeted strand. And in the next replication cycle, the ‘T:A’ base pair is repaired to ‘C:G’.