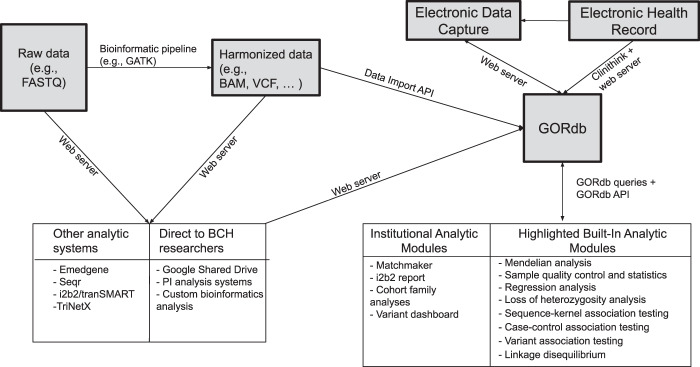
Fig. 3. Data flow diagram of genomics learning system.
Raw data is processed by secondary pipelines into harmonized data which is ingested into GORdb by the data import API. Phenotypic data from the EDC and EHR are also incorporated. Built-in GORdb queries, as well as institutionally-developed queries operate on the merged data, and can be executed by calling GORdb API or through the WuXi NextCODE user interface. Raw and harmonized data are also made available to other analytic systems and BCH researchers. Information from these systems are fed back into GORdb. Aspects of the GLS are connected by a Python web server, which executes data transfer to/from the GLS components, sends automated alerts to researchers about new data availability and warnings to bioinformaticians about potential metadata errors (for instance, duplicate subject enrollment).