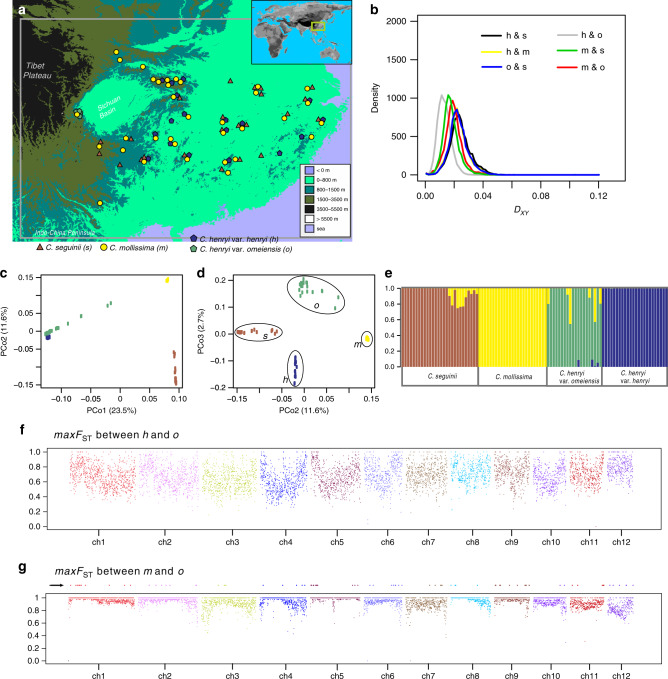
Fig. 1. The four Castanea taxa show distinct genomic variation.
a Map showing the distributions of samples collected. Each of the four taxa is shown in a different color. b Frequency polygon of DXY per base pair for each pair out of the four taxa, using a sliding-window approach with a window size of 100 kbp. Abbreviations are: s = C. seguinii, m = C. mollissima, h = C. henryi var. henryi, o = C. henryi var. omeiensis (the same below). c, d PCoA showing the first three coordinates and the percentage of variation explained by each coordinate. The four colors represent samples collected from the four taxa. e Population structure estimated by admixture analysis for all four Castanea taxa. f maxFST analysis of 100 kbp nonoverlapping windows across the 12 chromosomes between C. henryi var. henryi and C. henryi var. omeiensis. The 12 chromosomes are represented by different colors. g maxFST analysis of 100-kb nonoverlapping windows across the 12 chromosomes between C. mollissima and C. henryi var. omeiensis. The arrow in g indicates windows where fixed differences between C. henryi var. omeiensis and the two parental taxa (C. henryi var. henryi and C. mollissima) can be observed. Source data underlying Fig. 1b–g are provided as a Source data file.