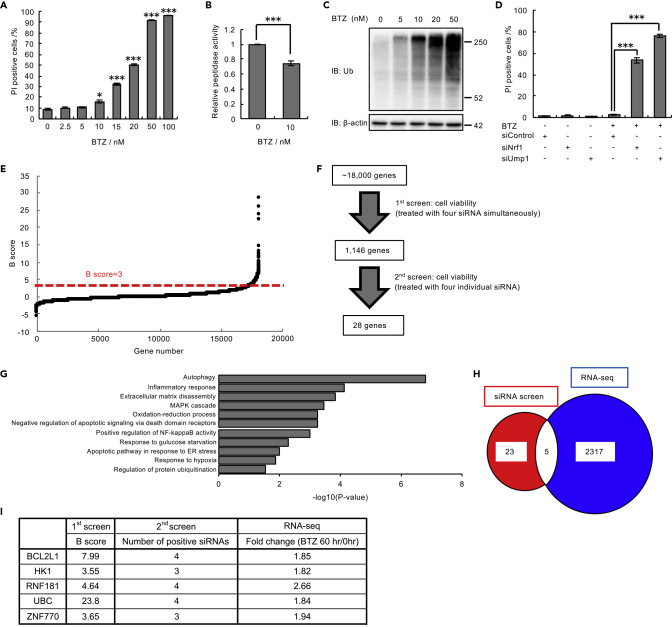
Figure 1.
Genome-wide siRNA Screen for Cell Death Suppressors under Proteasome Inhibition
(A) Viability assay of U2OS cells treated with bortezomib (BTZ) (0–100 nM) for 48 h.
(B) Proteasome peptidase activity of lysates from U2OS cells treated with 10 nM BTZ for 48 h.
(C) Immunoblot analysis of lysates from U2OS cells with antibodies against the indicated proteins. U2OS cells were treated with BTZ (0–50 nM) for 16 h before harvest.
(D) Viability assay of U2OS cells transfected with Nrf1 and Ump1 siRNAs or control siRNA and then treated with 10 nM BTZ for 48 h.
(E) B score of all samples in the first genome-wide siRNA screen. The results are ordered from lowest to highest. The dashed red line shows the cutoff value of positive hits in the first screen.
(F) The workflow of siRNA screens. In the first screen, U2OS cells were treated with a pooled siRNA library containing four siRNA constructs per gene. In the second screen, U2OS cells were treated with four individual siRNAs.
(G) Enrichment analysis of 2,322 genes upregulated by BTZ treatment for 60 h was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The gene expression values were log2 transformed, and GO analysis was performed. Representative data are shown.
(H) Venn diagram showing positive hits from the siRNA screens (red) and genes that were upregulated by BTZ treatment (blue). Five genes were common to the results of both screens (white).
(I) The final list of five hits and the scores of each assay in the two screens. The percentage of dead cells was assessed by FACS after staining with Hoechst 33,342 and propidium iodide (PI) in (A) and (D). Data in (A), (B), and (D) are the mean ± SEM (n = 3). Data in (A) and (B) were analyzed by two-tailed Student's t test (∗p < 0.05, ∗∗∗p < 0.001). Data in (D) were analyzed by two-way ANOVA followed by Tukey's test (∗∗∗p < 0.001). See also Figure S1 and Table S1.