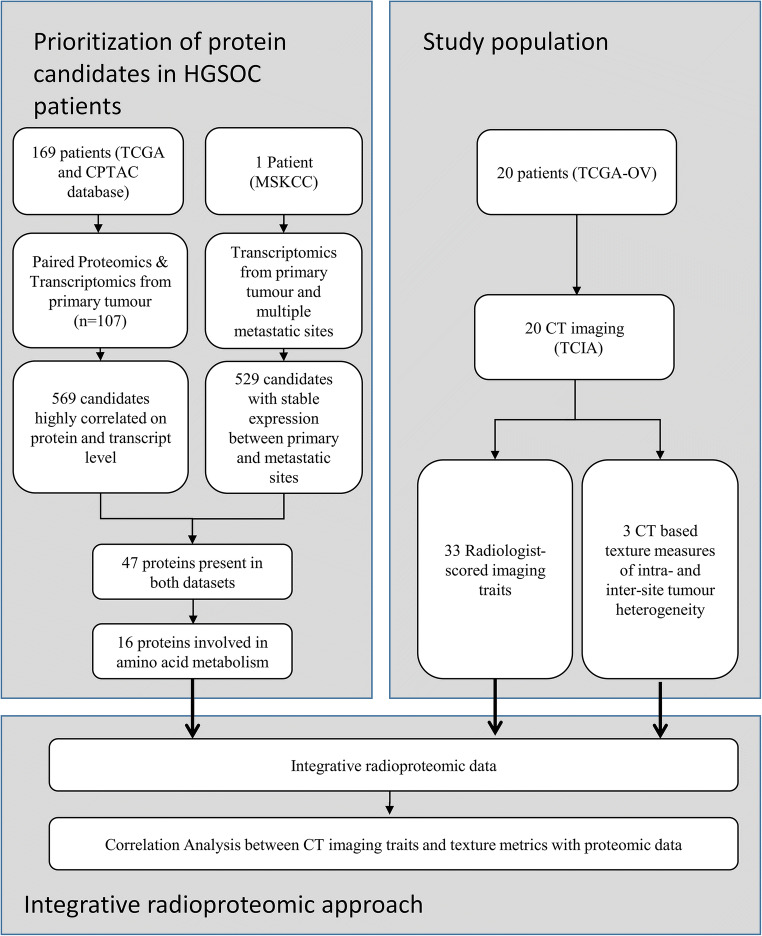
Fig. 1.
Study flow chart. Patients analyzed in this study were part of the TCGA dataset (n = 169). Protein candidates (5504 total) that exhibited stable expression between primary and metastatic sites and those that highly correlated with transcript abundance (n = 107 HGSOC patients, TCGA, and CPTAC) were selected using the Student’s t test (p value < 0.1) after boxCox normalization of transcript expression coefficients of variation and Spearman correlation distributions, respectively. Forty-seven genes exhibited both low correlation of variables between primary and metastatic sites (529 candidates) and were highly correlated at the protein and transcript levels (569 candidates). Among those, 16 proteins were associated with amino acid metabolism and selected for final analysis. CT data were available in 20 patients. Thirty-three radiologist-scored imaging traits and three CT-based texture measures of inter-site tumor heterogeneity were assessed and their correlation with the 16 cancer proteins was calculated