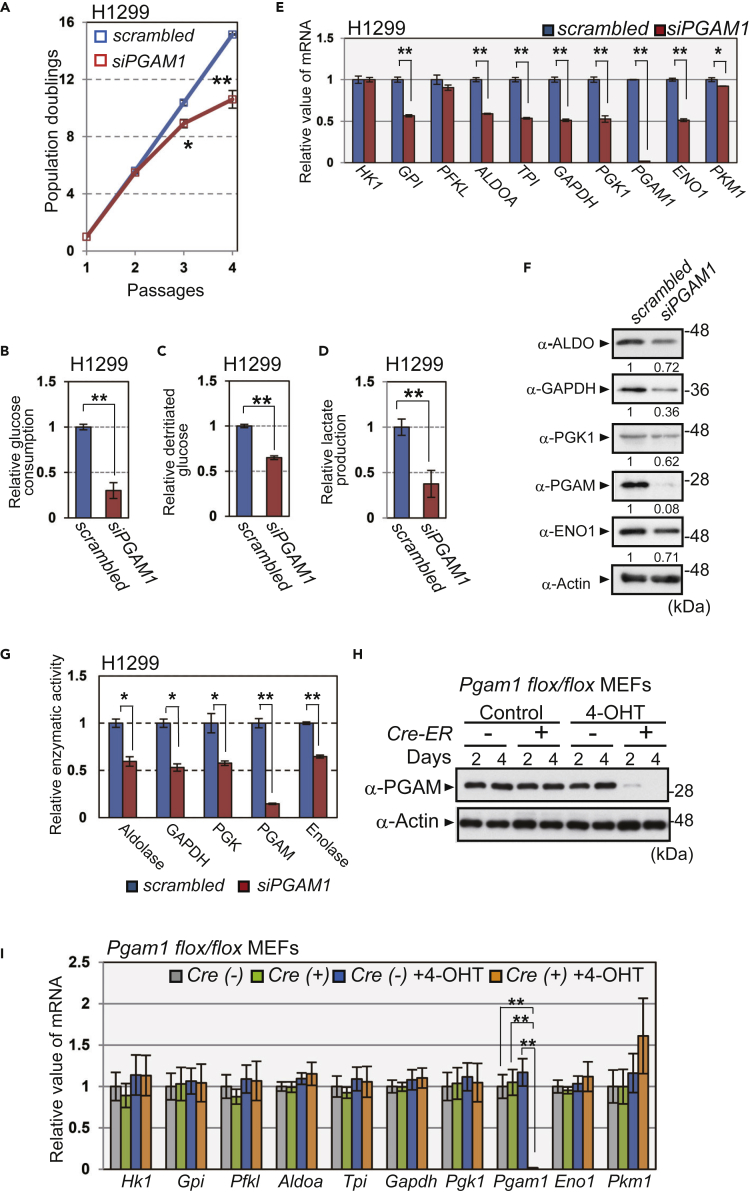
Figure 2.
Ablation of PGAM Downregulates Glycolysis In Vitro
The impact of PGAM1 knockdown on glycolysis in cancer cells in vitro. H1299 cells were transfected with siPGAM1 (n = 3) or scrambled RNA (n = 3) (A–G).
(A) PGAM1-knockdown H1299 cells were passaged according to 3T3 cell culture protocol. The proliferation curves show population doublings. The glycolytic profiles were evaluated in PGAM1-knockdown H1299 cells (B–G).
(B–D) Glucose consumption was evaluated by measuring glucose concentration in medium (B), whereas glycolytic flux was evaluated using 3H-labeled glucose (C). Lactate production was determined by the measurements of lactate concentration in medium (D).
(E) The mRNA levels for glycolytic enzymes were assessed. Data are relative to those in scrambled RNA-transfected H1299 cells. ∗p < 0.05 and ∗∗p < 0.005, Student's t test.
(F) The protein levels of the indicated glycolytic enzymes were analyzed by immunoblot using anti-aldolase (ALDO), anti-GAPDH, anti-PGK1, anti-PGAM, anti-enolase1 (ENO1), and anti-actin antibodies. Intensity of immunoblotting bands was normalized to that of actin. Relative values were shown, compared with scrambled RNA-transfected H1299 cells.
(G) Enzymatic activity of aldolase, GAPDH, PGK, PGAM, and enolase was measured by spectrometric assay.
(H and I) Pgam1 was ablated in primary MEFs from Pgam1flox/flox mice harboring Cre-ER by treatment with 4-hydroxytamoxifen (4-OHT) for 4 days. (H) PGAM protein levels in the indicated MEFs were analyzed by immunoblotting. (I) Comparison of glycolytic mRNAs among the indicated MEFs after 4-OHT treatment (n = 3). Cre(−); Pgam1flox/flox MEFs, Cre(+); Pgam1flox/flox + Cre-ER MEFs.
Data are relative to those in control. ∗p < 0.05 and ∗∗p < 0.005, Dunnett's multiple comparison test. Data represent the mean ± SEM.
See also Figures S3 and S4.