Version Changes
Revised. Amendments from Version 2
A typo in the equation on p 80% was corrected; originally it was 1− p 80%=1−∫NB(⌊ x⌋; k, k/( R 0+ k))d x, which should have been instead 1− p 80%=∫NB(⌊ x⌋; k, k/( R 0+ k))d x. The typo was only present in the manuscript and did not affect the analysis or other parts of the manuscript.
Abstract
Background: A novel coronavirus disease (COVID-19) outbreak has now spread to a number of countries worldwide. While sustained transmission chains of human-to-human transmission suggest high basic reproduction number R 0, variation in the number of secondary transmissions (often characterised by so-called superspreading events) may be large as some countries have observed fewer local transmissions than others.
Methods: We quantified individual-level variation in COVID-19 transmission by applying a mathematical model to observed outbreak sizes in affected countries. We extracted the number of imported and local cases in the affected countries from the World Health Organization situation report and applied a branching process model where the number of secondary transmissions was assumed to follow a negative-binomial distribution.
Results: Our model suggested a high degree of individual-level variation in the transmission of COVID-19. Within the current consensus range of R 0 (2-3), the overdispersion parameter k of a negative-binomial distribution was estimated to be around 0.1 (median estimate 0.1; 95% CrI: 0.05-0.2 for R0 = 2.5), suggesting that 80% of secondary transmissions may have been caused by a small fraction of infectious individuals (~10%). A joint estimation yielded likely ranges for R 0 and k (95% CrIs: R 0 1.4-12; k 0.04-0.2); however, the upper bound of R 0 was not well informed by the model and data, which did not notably differ from that of the prior distribution.
Conclusions: Our finding of a highly-overdispersed offspring distribution highlights a potential benefit to focusing intervention efforts on superspreading. As most infected individuals do not contribute to the expansion of an epidemic, the effective reproduction number could be drastically reduced by preventing relatively rare superspreading events.
Keywords: COVID-19, SARS-CoV-2, novel coronavirus, overdispersion, superspreading, branching process
Introduction
A novel coronavirus disease (COVID-19) outbreak, which is considered to be associated with a market in Wuhan, China, is now affecting a number of countries worldwide 1, 2. A substantial number of human-to-human transmission has occurred; the basic reproduction number R 0 (the average number of secondary transmissions caused by a single primary case in a fully susceptible population) has been estimated around 2–3 3– 5. More than 100 countries have observed confirmed cases of COVID-19. A few countries have already been shifting from the containment phase to the mitigation phase 6, 7, with a substantial number of locally acquired cases (including those whose epidemiological link is untraceable). On the other hand, there are countries where a number of imported cases were ascertained but fewer secondary cases have been reported than might be expected with an estimated value of R 0 of 2–3.
This suggests that not all symptomatic cases cause a secondary transmission, which was also estimated to be the case for past coronavirus outbreaks (SARS/MERS) 8, 9. High individual-level variation (i.e. overdispersion) in the distribution of the number of secondary transmissions, which can lead to so-called superspreading events, is crucial information for epidemic control 9. High variation in the distribution of secondary cases suggests that most cases do not contribute to the expansion of the epidemic, which means that containment efforts that can prevent superspreading events have a disproportionate effect on the reduction of transmission.
We estimated the level of overdispersion in COVID-19 transmission by using a mathematical model that is characterised by R 0 and the overdispersion parameter k of a negative binomial branching process. We fit this model to worldwide data on COVID-19 cases to estimate k given the reported range of R 0 and interpret this in the context of superspreading.
Methods
Data source
We extracted the number of imported/local cases in the affected countries ( Table 1) from the WHO situation report 38 10 published on 27 February 2020, which was the latest report of the number of imported/local cases in each country (as of the situation report 39, WHO no longer reports the number of cases stratified by the site of infection). As in the WHO situation reports, we defined imported cases as those whose likely site of infection is outside the reporting country and local cases as those whose likely site of infection is inside the reporting country. Those whose site of infection was under investigation were excluded from the analysis (Estonia had no case with a known site of infection and was excluded). In Egypt and Iran, no imported cases have been confirmed, which cause the likelihood value to be zero; data in these two countries were excluded. To distinguish between countries with and without an ongoing outbreak, we extracted daily case counts from an online resource 11 and determined the dates of the latest case confirmation for each country (as of 27 February).
Table 1. The number of confirmed COVID-19 cases reported (as of 27 February 2020).
| Country | Total
cases |
Imported
cases |
Local
cases |
Site of infection
unknown |
Deaths | Latest date of case
confirmation |
|---|---|---|---|---|---|---|
| South Korea | 1766 | 17 | 605 | 1144 | 13 | 27/02/2020 |
| Japan | 186 | 39 | 129 | 18 | 3 | 27/02/2020 |
| Singapore | 93 | 24 | 69 | 0 | 0 | 27/02/2020 |
| Australia | 23 | 20 | 3 | 0 | 0 | 26/02/2020 |
| Malaysia | 22 | 20 | 2 | 0 | 0 | 27/02/2020 |
| Vietnam * | 16 | 8 | 8 | 0 | 0 | 13/02/2020 |
| Philippines * | 3 | 3 | 0 | 0 | 1 | 05/02/2020 |
| Cambodia * | 1 | 1 | 0 | 0 | 0 | 30/01/2020 |
| Thailand | 40 | 23 | 7 | 10 | 0 | 26/02/2020 |
| India * | 3 | 3 | 0 | 0 | 0 | 03/02/2020 |
| Nepal * | 1 | 1 | 0 | 0 | 0 | 24/01/2020 |
| Sri Lanka | 1 | 1 | 0 | 0 | 0 | 27/01/2020 |
| USA | 59 | 56 | 2 | 1 | 0 | 26/02/2020 |
| Canada | 11 | 9 | 1 | 1 | 0 | 27/02/2020 |
| Brazil | 1 | 1 | 0 | 0 | 0 | 26/02/2020 |
| Italy | 400 | 3 | 121 | 276 | 12 | 27/02/2020 |
| Germany | 21 | 3 | 14 | 4 | 0 | 27/02/2020 |
| France | 18 | 8 | 7 | 3 | 2 | 27/02/2020 |
| UK | 13 | 12 | 1 | 0 | 0 | 27/02/2020 |
| Spain | 12 | 10 | 1 | 1 | 0 | 27/02/2020 |
| Croatia | 3 | 2 | 1 | 0 | 0 | 26/02/2020 |
| Austria | 2 | 2 | 0 | 0 | 0 | 27/02/2020 |
| Finland | 2 | 2 | 0 | 0 | 0 | 26/02/2020 |
| Israel | 2 | 2 | 0 | 0 | 0 | 27/02/2020 |
| Russia * | 2 | 2 | 0 | 0 | 0 | 31/01/2020 |
| Sweden | 2 | 2 | 0 | 0 | 0 | 27/02/2020 |
| Belgium * | 1 | 1 | 0 | 0 | 0 | 04/02/2020 |
| Denmark | 1 | 1 | 0 | 0 | 0 | 27/02/2020 |
| Estonia † | 1 | 0 | 0 | 1 | 0 | 27/02/2020 |
| Georgia | 1 | 1 | 0 | 0 | 0 | 26/02/2020 |
| Greece | 1 | 1 | 0 | 0 | 0 | 27/02/2020 |
| North Macedonia | 1 | 1 | 0 | 0 | 0 | 26/02/2020 |
| Norway | 1 | 1 | 0 | 0 | 0 | 27/02/2020 |
| Romania | 1 | 1 | 0 | 0 | 0 | 26/02/2020 |
| Switzerland | 1 | 1 | 0 | 0 | 0 | 27/02/2020 |
| Iran † | 141 | 0 | 28 | 113 | 22 | 27/02/2020 |
| Kuwait | 43 | 43 | 0 | 0 | 0 | 27/02/2020 |
| Bahrain | 33 | 33 | 0 | 0 | 0 | 26/02/2020 |
| UAE | 13 | 8 | 5 | 0 | 0 | 27/02/2020 |
| Iraq | 6 | 6 | 0 | 0 | 0 | 27/02/2020 |
| Oman | 4 | 4 | 0 | 0 | 0 | 27/02/2020 |
| Lebanon | 1 | 1 | 0 | 0 | 0 | 27/02/2020 |
| Pakistan | 2 | 1 | 0 | 1 | 0 | 26/02/2020 |
| Afghanistan | 1 | 1 | 0 | 0 | 0 | 24/02/2020 |
| Egypt * † | 1 | 0 | 1 | 0 | 0 | 14/02/2020 |
| Algeria | 1 | 1 | 0 | 0 | 0 | 25/02/2020 |
* Countries considered to be without an ongoing outbreak
† Countries excluded from analysis
Model
Assuming that the offspring distributions (distribution of the number of secondary transmissions) for COVID-19 cases are identically- and independently-distributed negative-binomial distributions, we constructed the likelihood of observing the reported number of imported/local cases (outbreak size) of COVID-19 for each country. The probability mass function for the final cluster size resulting from s initial cases is, according to Blumberg et al. 12, given by
If the observed case counts are part of an ongoing outbreak in a country, cluster sizes may grow in the future. To address this issue, we adjusted the likelihood for those countries with ongoing outbreak by only using the condition that the final cluster size of such a country has to be larger than the currently observed number of cases. The corresponding likelihood function is
with a convention We assumed that the growth of a cluster in a country had ceased if 7 days have passed since the latest reported case (denoted by set A). We applied the final size likelihood c( x; s) to those countries and c o( x; s) to the rest of the countries (countries with an ongoing outbreak: B). The total likelihood is
Statistical analysis
Varying the assumed R 0 between 0–5 (fixed at an evenly-spaced grid of values), we estimated the overdispersion parameter k using the likelihood function described above. We used the Markov-chain Monte Carlo (MCMC) method to provide 95% credible intervals (CrIs). The reciprocal of k was sampled where the prior distribution for the reciprocal was weakly-informed half-normal (HalfNormal( σ = 10)). We employed the adaptive hit-and-run Metropolis algorithm 13 and obtained 500 thinned samples from 10,000 MCMC steps (where the first half of the chain was discarded as burn-in). We confirmed that the final 500 samples have an effective sample size of at least 300, indicating sufficiently low auto-correlation.
We also performed a joint-estimation of R 0 and k by the MCMC method (with a weakly-informed normal prior N( μ = 3, σ = 5) for R 0 and the weakly-informed half-normal prior (HalfNormal( σ = 10)) for the reciprocal of k.
Statistical analysis was implemented in R-3.6.1 with a package { LaplacesDemon}-16.1.1. The reproducible code for this study is available on GitHub 14.
Proportion responsible for 80% of secondary transmissions
Using the estimated R 0 and k, we computed the estimated proportion of infected individuals responsible for 80% of the total secondary transmissions. Such proportion p 80% is given as
where X satisfies
Here, represents the probability mass of a negative-binomial distribution with a mean R 0 and an overdispersion parameter k. This calculation is eased by the following rearrangement:
We computed p 80% for each MCMC (Markov-chain Monte Carlo) sample to yield median and 95% CrIs.
Model comparison with a Poisson branching process model
To test if our assumption of overdispersed offspring distribution better describes the data, we compared our negative-binomial branching process model with a Poisson branching process model, which assumes that the offspring distribution follows a Poisson distribution instead of negative-binomial. Since a negative-binomial distribution converges to a Poisson distribution as k → ∞, we approximately implemented a Poisson branching process model by fixing k of the negative-binomial model at 10 10. We compared the two models by the widely-applicable Bayesian information criterion (WBIC) 15.
Simulation of the effect of underreporting
We used simulations to investigate potential bias caused by underreporting, one of the major limitations of the present study. Underreporting in some countries may be more frequent than others because of limited surveillance and/or testing capacity, causing heterogeneity in the number of cases that could have affected the estimated overdispersion. See Extended data (Supplementary materials) 16 for detailed methods.
The effect of a differential reproduction number for imported cases
Due to interventions targeting travellers (e.g. screening and quarantine), the risk of transmission from imported cases may be lower than that from local cases. As part of the sensitivity analysis in Extended data, we estimated k assuming that the reproduction number of imported cases is smaller than that of local cases.
Results
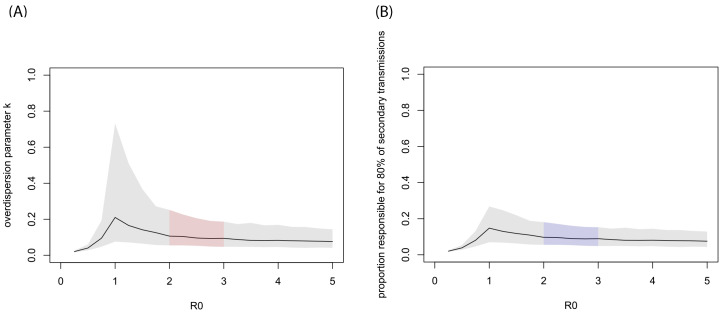
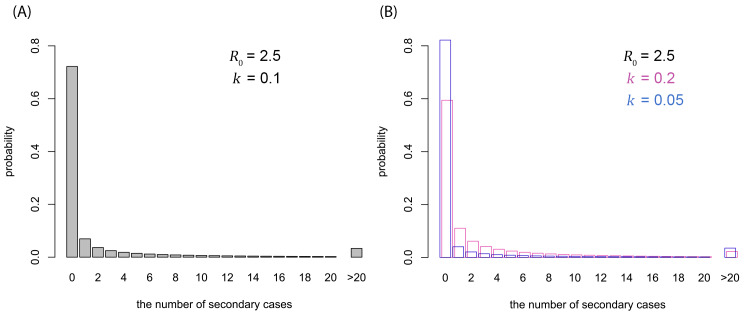
Our estimation suggested substantial overdispersion ( k ≪ 1) in the offspring distribution of COVID-19 ( Figure 1A and Figure 2). Within the current consensus range of R 0 (2–3), k was estimated to be around 0.1 (median estimate 0.1; 95% CrI: 0.05–0.2 for R 0 = 2.5). For the R 0 values of 2–3, the estimates suggested that 80% of secondary transmissions may have been caused by a small fraction of infectious individuals (~10%; Figure 1B).
Figure 1. MCMC estimates given assumed R 0 values.
( A) Estimated overdispersion parameter for various basic reproduction number R 0. ( B) The proportion of infected individuals responsible for 80% of the total secondary transmissions ( p 80%). The black lines show the median estimates given fixed R 0 values and the grey shaded areas indicate 95% CrIs. The regions corresponding to the likely range of R 0 (2–3) are indicated by colour.
Figure 2. Possible offspring distributions of COVID-19.
( A) Offspring distribution corresponding to R 0 = 2.5 and k = 0.1 (median estimate). ( B) Offspring distribution corresponding to R 0 = 2.5 and k = 0.05 (95% CrI lower bound), 0.2 (upper bound). The probability mass functions of negative-binomial distributions are shown.
The result of the joint estimation suggested the likely bounds for R 0 and k (95% CrIs: R 0 1.4–12; k 0.04–0.2). The upper bound of R 0 did not notably differ from that of the prior distribution (=13.5), suggesting that our model and the data only informed the lower bound of R 0. This was presumably because the contribution of R 0 to the shape of a negative-binomial distribution is marginal when k is small ( Extended data, Figure S1) 16. A scatterplot ( Extended data, Figure S2) 16 exhibited a moderate correlation between R 0 and k (correlation coefficient -0.4).
Model comparison between negative-binomial and Poisson branching process models suggested that a negative-binomial model better describes the observed data; WBIC strongly supported the negative-binomial model with a difference of 11.0 ( Table 2). The simulation of the effect of underreporting suggested that possible underreporting is unlikely to cause underestimation of overdispersion parameter k ( Extended data, Figure S3) 16. A slight increase in the estimate of k was observed when the reproduction number for imported cases was assumed to be lower due to interventions ( Extended data, Table S1).
Table 2. Model comparison between negative-binomial and Poisson branching process models.
| Model | Parameter 95% CrIs | WBIC | ΔWBIC | |
|---|---|---|---|---|
| R 0 | k | |||
| Negative-binomial | 1.4–12 | 0.04-0.2 | 45.6 | 0 |
| Poisson | 0.95–1.2 | 10 10 (fixed) | 56.6 | 11.0 |
Discussion
Our results suggested that the offspring distribution of COVID-19 is highly overdispersed. For the likely range of R 0 of 2–3, the overdispersion parameter k was estimated to be around 0.1, suggesting that the majority of secondary transmission may be caused by a very small fraction of individuals (80% of transmissions caused by ~10% of the total cases). These results are consistent with a number of observed superspreading events observed in the current COVID-19 outbreak 17, and also in line with the estimates from the previous SARS/MERS outbreaks 8.
The overdispersion parameter for the current COVID-19 outbreak has also been estimated by stochastic simulation 18 and from contact tracing data in Shenzhen, China 19. The former study did not yield an interpretable estimate of k due to the limited data input. In the latter study, the estimates of R e (the effective reproduction number) and k were 0.4 (95% confidence interval: 0.3–0.5) and 0.58 (0.35–1.18), respectively, which did not agree with our findings. However, these estimates were obtained from pairs of cases with a clear epidemiological link and therefore may have been biased (downward for R 0 and upward for k) if superspreading events had been more likely to be missed during the contact tracing.
Although cluster size distributions based on a branching process model are useful in inference of the offspring distribution from limited data 12, 20, they are not directly applicable to an ongoing outbreak because the final cluster size may not yet have been observed. In our analysis, we adopted an alternative approach which accounts for possible future growth of clusters to minimise the risk of underestimation. As of 27 February 2020, the majority of the countries in the dataset had ongoing outbreaks (36 out of 43 countries analysed, accounting for 2,788 cases of the total 2,816). Even though we used the case counts in those countries only as the lower bounds of future final cluster sizes, which might have only partially informed of the underlying branching process, our model yielded estimates with moderate uncertainty levels (at least sufficient to suggest that k may be below 1). Together with the previous finding suggesting that the overdispersion parameter is unlikely to be biased downwards 21, we believe our analysis supports the possibility of highly-overdispersed transmission of COVID-19.
A number of limitations need to be noted in this study. We used the confirmed case counts reported to WHO and did not account for possible underreporting of cases. Heterogeneities between countries in surveillance and intervention capacities, which might also be contributing to the estimated overdispersion, were not considered (although we investigated such effects by simulations; see Extended data, Figure S3) 16. Reported cases whose site of infection classified as unknown, which should in principle be counted as either imported or local cases, were excluded from analysis. Some cases with a known site of infection could also have been misclassified (e.g., cases with travel history may have been infected locally). The distinction between countries with and without ongoing outbreak (7 days without any new confirmation of cases) was arbitrary. However, we believe that our conclusion is robust because the distinction does not change with different thresholds (4–14 days), within which the serial interval of SARS-CoV-2 is likely to fall 22, 23.
Our finding of a highly-overdispersed offspring distribution suggests that there is benefit to focusing intervention efforts on superspreading. As most infected individuals do not contribute to the expansion of transmission, the effective reproduction number could be drastically reduced by preventing relatively rare superspreading events. Identifying characteristics of settings that could lead to superspreading events will play a key role in designing effective control strategies.
Data availability
Source data
Zenodo: Extended data: Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. https://doi.org/10.5281/zenodo.3740348 16.
This project contains the following source data taken from references 10 and 11:
Extended data
Zenodo: Extended data: Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. https://doi.org/10.5281/zenodo.3911576 16.
This project contains the following extended data
supplementarymaterials.pdf. (Supplementary material: Estimating the amount of superspreading using outbreak sizes of COVID-19 outside China.)
figS1.tif. (Figure S1. Offspring distributions for different R 0 values. The probability mass functions of negativebinomial distributions are shown. The overdispersion parameter k is fixed at 0.1.)
figS2.tif. (Supplementary Figure 2. Scatter plot of MCMC samples from a joint estimation of R 0 and k. The dotted line represents the threshold R 0 = 1)
figS3.tif. (Supplementary Figure 3. Estimates of overdispersion from simulations with underreporting. (A) Maximum-likelihood estimates (MLEs) of overdispersion parameter k with different distributions for country-specific reporting probability q i (including constant q i = 1). Both imported and local cases are assumed to be reported at probability q i in country i. The blue dotted line indicates the true value k = 0.1. (B) MLEs where imported cases were assumed to be fully reported and local cases were reported at probability q i. (C) Probability density functions for beta distributions used in the simulation.)
Code availability
The reproducible code is available at: https://github.com/akira-endo/COVID19_clustersize.
Archived code at time of publication: https://doi.org/10.5281/zenodo.3741743 14.
License: MIT.
Acknowledgements
This study was greatly motivated and inspired by the analysis published online by Kyra Grantz, C. Jessica E. Metcalf and Justin Lessler ( https://hopkinsidd.github.io/nCoV-Sandbox/DispersionExploration.html). We thank the authors for insightful inputs and contribution. We also thank Seth Blumberg for valuable feedback.
Members of the Centre for Mathematical Modelling of Infectious Diseases (CMMID) COVID-19 Working Group (random order):
Rosalind M Eggo, Billy J Quilty, Nikos I Bosse, Kevin van Zandvoort, James D Munday, Stefan Flasche, Alicia Rosello, Mark Jit, W John Edmunds, Amy Gimma, Yang Liu, Kiesha Prem, Hamish Gibbs, Charlie Diamond, Christopher I Jarvis, Nicholas Davies, Fiona Sun, Joel Hellewell, Timothy W Russell, Thibaut Jombart, Samuel Clifford, Petra Klepac, Graham Medley, Carl A B Pearson
CMMID COVID-19 working group funding statements:
Rosalind M Eggo (HDR UK (MR/S003975/1)), Billy J Quilty (National Institute for Health Research (NIHR) (16/137/109)), Kevin van Zandvoort (Elrha’s Research for Health in Humanitarian Crises (R2HC) Programme), James D Munday (Wellcome Trust (210758/Z/18/Z)), Stefan Flasche (Wellcome Trust (208812/Z/17/Z)), Alicia Rosello (NIHR (PR-OD-1017-20002)), Mark Jit (Gates (INV-003174), NIHR (16/137/109)), Amy Gimma (RCUK/ ESRC (ES/P010873/1)), Yang Liu (Gates (INV-003174), NIHR (16/137/109)), Kiesha Prem (Gates (INV-003174)), Hamish Gibbs (NIHR (ITCRZ 03010)), Charlie Diamond (NIHR (16/137/109)), Christopher I Jarvis (RCUK/ESRC (ES/P010873/1)), Nicholas Davies (NIHR (HPRU-2012-10096)), Fiona Sun (NIHR EPIC grant (16/137/109)), Joel Hellewell (Wellcome Trust (210758/Z/18/Z)), Timothy W Russell (Wellcome Trust (206250/Z/17/Z)), Thibaut Jombart (RCUK/ESRC (ES/P010873/1), UK PH RST, NIHR HPRU Modelling Methodology), Samuel Clifford (Wellcome Trust (208812/Z/17/Z)), Petra Klepac (Gates (INV-003174))
Funding Statement
SA [210758], AJK [206250] and SF [210758] are supported by the Wellcome Trust. AE is financially supported by The Nakajima Foundation and The Alan Turing Institute.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 3; peer review: 2 approved]
References
- 1. Zhu N, Zhang D, Wang W, et al. : A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lai CC, Shih TP, Ko WC, et al. : Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3): 105924. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao S, Lin Q, Ran J, et al. : Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. 10.1016/j.ijid.2020.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang S, Diao M, Yu W, et al. : Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: A data-driven analysis. Int J Infect Dis. 2020;93:201–204. 10.1016/j.ijid.2020.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbott S, Hellewell J, Munday J, et al. : The transmissibility of novel Coronavirus in the early stages of the 2019-20 outbreak in Wuhan: Exploring initial point-source exposure sizes and durations using scenario analysis [version 1; peer review: 1 approved]. Wellcome Open Res. 2020;5:17. 10.12688/wellcomeopenres.15718.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Headquarters for Novel Coronavirus Disease Control; Ministry of Health Labour and Welfare: Basic Policies for Novel Coronavirus Disease Control.2020. Reference Source [Google Scholar]
- 7. Department of Health and Social Care, Hancock M: Press release: Government outlines further plans to support health and social care system in fight against COVID-19.2020; [cited 9 Mar 2020]. Reference Source [Google Scholar]
- 8. Kucharski AJ, Althaus CL: The role of superspreading in Middle East respiratory syndrome coronavirus (MERS-CoV) transmission. Euro Surveill. 2015;20(25):14–8. 10.2807/1560-7917.es2015.20.25.21167 [DOI] [PubMed] [Google Scholar]
- 9. Lloyd-Smith JO, Schreiber SJ, Kopp PE, et al. : Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438(7066):355–359. 10.1038/nature04153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. World Health Organization: Coronavirus disease 2019 (COVID-19) Situation Report – 38.2020. Reference Source [Google Scholar]
- 11. COVID2019.app - LIVE stats and graphs.2020; [cited 4 Mar 2020]. Reference Source [Google Scholar]
- 12. Blumberg S, Funk S, Pulliam JR: Detecting differential transmissibilities that affect the size of self-limited outbreaks. Wilke CO, editor. PLoS Pathog. 2014;10(10):e1004452. 10.1371/journal.ppat.1004452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen MH, Schmeiser B: Performance of the Gibbs, Hit-and-Run, and Metropolis Samplers. J Comput Graph Stat. 1993;2(3):251–272. 10.2307/1390645 [DOI] [Google Scholar]
- 14. Endo A, Abbott S, Kucharski AJ, et al. : Estimating the amount of superspreading using outbreak sizes of COVID-19 outside China (Version v1.0.0). Zenodo. 2020. 10.5281/zenodo.3741743 [DOI] [Google Scholar]
- 15. Watanabe S: A Widely Applicable Bayesian Information Criterion.2013;14:867–897. 10.1088/0953-8984/23/18/184115 [DOI] [Google Scholar]
- 16. Endo A, Abbott S, Kucharski AJ, et al. : Extended data: Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Zenodo. 2020. 10.5281/zenodo.3911576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y, Eggo RM, Kucharski AJ: Secondary attack rate and superspreading events for SARS-CoV-2. Lancet. 2020;395(10227):e47. 10.1016/S0140-6736(20)30462-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riou J, Althaus CL: Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020;25(4): 2000058. 10.2807/1560-7917.ES.2020.25.4.2000058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bi Q, Wu Y, Mei S, et al. : Epidemiology and Transmission of COVID-19 in Shenzhen China: Analysis of 391 cases and 1,286 of their close contacts. medRxiv. 2020; 2020.03.03.20028423. 10.1101/2020.03.03.20028423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blumberg S, Lloyd-Smith JO: Inference of R(0) and transmission heterogeneity from the size distribution of stuttering chains.Ferguson N, editor. PLoS Comput Biol. 2013;9(5):e1002993. 10.1371/journal.pcbi.1002993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lloyd-Smith JO: Maximum Likelihood Estimation of the Negative Binomial Dispersion Parameter for Highly Overdispersed Data, with Applications to Infectious Diseases. Rees M, editor. PLoS One. 2007;2(2):e180. 10.1371/journal.pone.0000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Li Q, Guan X, Wu P, et al. : Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199–1207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishiura H, Linton NM, Akhmetzhanov AR: Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. 2020;93:284–286. 10.1016/j.ijid.2020.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]