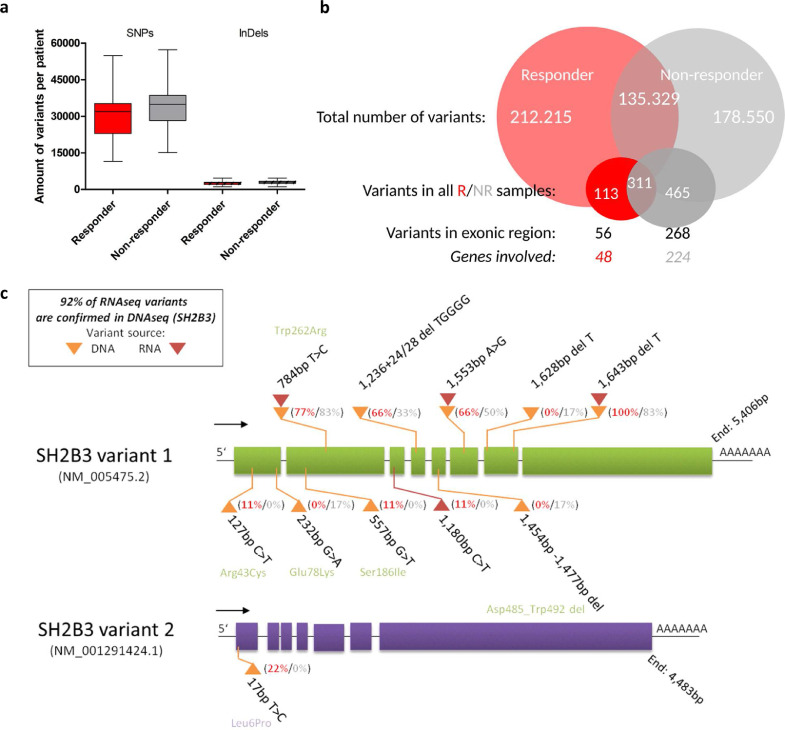
Fig. 4.
Summary of genetic mutation signature analysis in PERFECT patients via sequencing analysis. a: Transcriptomic variants identified through RNA-Seq data analysis. Plot shows the average number of variants (SNPs and InDels) per patient that have been identified by applying our customized transcriptomic variant calling pipeline and filtering approaches. SNPs and InDels are considered as successfully called, if at least five independent reads support the individual variant. b: Venn diagram for the RvsNR variant comparison, exonic region association, and unique gene identification. c: Targeted DNA-Seq (yellow triangle) and RNA-Seq (red triangle) variant summary of SH2B3. The plot shows the ratio of SNP/del sites that are identified in Responders (red) and Non-responders (grey) as well as the possible amino acid transfer from its origin to its potential replacement (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).