Fig. 5.
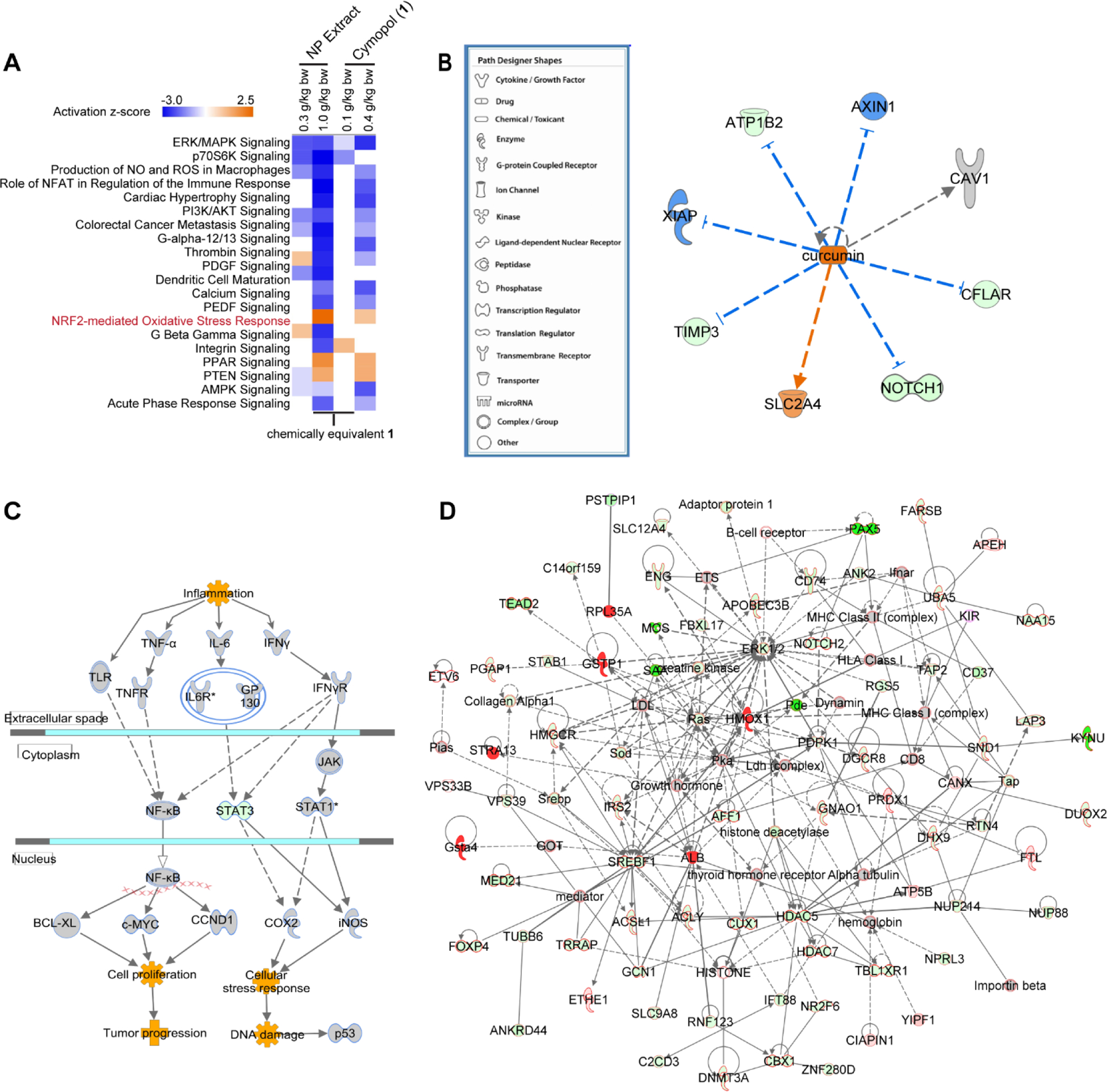
RNA-seq data analysis of mouse large intestines. (A-D) IPA analysis on RNA-seq data (n = 3) from mice large intestines. (A) Top canonical pathways derived from comparison analysis of all doses/treatments (B) Similar directional changes of cymopol (1) to curcumin. The blue lines indicate genes which are downregulated by curcumin, whereas the orange lines are for genes which are up-regulated. (C) Colorectal Cancer Metastasis Signaling pathway indicates that proinflammatory and oncogenic genes such as Tnf-α, Il6r, iNOS, and c-Myc are all downregulated, while cytoprotective Stat1 is up-regulated. (D) Top cancer relevant pathways regulated by NP extract in mice large intestine, green fill = downregulated; red fill = upregulated; outline of black = cancer functionality, outline of blue = abdominal cancer functionality, outline of red = both general cancer and abdominal cancer functionalities. Nrf2 target genes, Gsta4, Hmox1, and Gstp1, are all notably upregulated.
