Abstract
Background:
Prostate cancer (PRCA) is an androgen-driven disease, where androgens act through the androgen receptor (AR) to induce proliferation and survival of tumor cells. Recently, AR splice variant 7 (ARv7) has been implicated in advanced stages of PRCA and clinical recurrence. With the widespread use of AR-targeted therapies, there has been a rising interest in the expression of full-length AR and ARv7 in PRCA progression and how these receptors, both independently and together, contribute to adverse clinicopathologic outcomes.
Methods:
Despite a multitude of studies measuring the expression levels of AR and ARv7 in PRCA progression, the results have been inconsistent and sometimes contradictory due to technical and analytical discrepancies. To circumvent these inconsistencies, we used an automated multiplexed immunostaining platform for full-length AR and ARv7 in human PRCA samples, and objectively quantified expression changes with machine-learning based software. With this technology, we can assess receptor prevalence both independently, and co-expressed, within specific tissue and cellular compartments.
Results:
Full-length AR and ARv7 expression increased in epithelial nuclei of metastatic samples compared to benign. Interestingly, a population of cells with undetectable AR persisted though all stages of PRCA progression. Co-expression analyses showed an increase of the double positive (AR+/ARv7+) population in metastases compared to benign, and an increase of the double negative population in PRCA samples compared to benign. Importantly, analysis of clinicopathologic outcomes associated with AR/ARv7 co-expression showed a significant decrease of the double positive population with higher Gleason score, as well as in samples with recurrence in under 5 years. Conversely, the double negative population was significantly increased in samples with higher Gleason score and in samples with recurrence in under 5 years.
Conclusions:
Changes in AR and ARv7 co-expression may have prognostic value in PRCA progression and recurrence. A better understanding of the prevalence and clinicopathologic outcomes associated with changes in these receptors’ co-expression may provide a foundation for improved diagnosis and therapy for men with PRCA.
Keywords: androgens, splice variants, recurrence, machine-learning, multispectral imaging
INTRODUCTION
Prostate cancer (PRCA) is an androgen-driven disease, where proliferation of epithelial cells occurs primarily through downstream signaling of the androgen receptor (AR)1. When AR is bound by ligand, it dimerizes and translocates to the nucleus, where it acts as a transcription factor for targets genes involved in growth and proliferation2. This androgen driven signaling pathway is the target for PRCA therapies (i.e. luteinizing hormone-releasing hormone agonists, androgen receptor antagonists) which reduce tumor burden and serum biomarker expression3. Despite initial success, however, androgen deprivation therapies inevitably fail, resulting in disease recurrence4. Importantly, our current knowledge of biomarkers for PRCAs that will progress to recurrence is inadequate. A thorough understanding of the androgen pathway in advanced disease and recurrence may allow improved diagnosis and treatment of patients that will develop recurrent disease.
There are several proposed models for the development of disease recurrence, including androgen-dependent and -independent mechanisms5. Androgen-dependent processes contributing to recurrence include AR amplification, intra-tumoral androgen synthesis, altered AR co-factor expression, or AR mutation leading to ligand promiscuity6–9. Alternatively, recurrence can occur through androgen-independent mechanisms including growth factor mediated AR signaling and the expansion of AR negative cell types (i.e. prostate cancer stem-like cells, neuroendocrine prostate cancer cells, double negative prostate cancer)10–14. Recent evidence has supported the emergence of AR splice variant 7 (ARv7) expression in advanced disease and recurrence15–17. ARv7 lacks the ligand-binding domain of full-length AR, and thus does not require ligand to translocate to the nucleus18. Interestingly, both AR and ARv7 require dimerization for nuclear translocation, and have been shown to interact as heterodimers that translocate to the nucleus together19, 20. This interaction between AR and ARv7 is not well understood.
Because the androgen pathway is critical for the proliferation, cell survival, and growth of prostate tumors, many studies have sought to identify the prevalence and significance of AR and ARv7 expression in PRCA progression with inconsistent results. It is generally recognized that transcriptionally active full-length AR is expressed in nuclei of both stromal and epithelial prostate cells; however, while some studies show AR expression increasing in PRCA progression and recurrence21–23, others show a decrease of receptor expression in advanced disease24–26. Similarly, while ARv7 expression has been shown to increase through PRCA progression, particularly in recurrence27, 28, little is known about ARv7 expression early in PRCA progression. This partial understanding of AR expression in PRCA could be due to: 1) variability of protocol procedures (i.e. fixation, antigen retrieval, chromogen), 2) subjective analysis techniques, 3) insufficient reagents such as a lack of full length AR-specific (AR C-terminal)/AR variant-specific (ARv7) antibodies, or 4) AR expression heterogeneity leading to analysis of hot spots for AR expression. Given the important role of AR and its variants in PRCA, it is imperative that we as a field assess the expression of these receptors objectively to accurately determine the incidence, stage specificity, and clinical relevance of AR and ARv7 expression, both independently and co-expressed.
In this study, we objectively quantified AR and ARv7 expression in human prostate samples using multiplexed immunohistochemistry (IHC), multispectral imaging, and machine-learning based analysis software. With this method, it is possible to quantify receptor co-expression providing insight into the potential interaction of these two receptors in PRCA progression, and its clinical significance. A better understanding of AR and ARv7 expression in PRCA progression may serve as a platform for better diagnosis and treatment in patients with PRCA.
MATERIALS AND METHODS
Prostate Tissue Microarrays (TMAs)
To assess AR and ARv7 protein localization and abundance during PRCA progression, we used a previously described “progression” tissue microarray (pTMA) composed of duplicate 6mm tissue cores from patients at varying stages of PRCA progression29, 30. Tissue cores consisted of benign prostate tissue (BPT; n=101 cores, 52 patients), high-grade prostatic intraepithelial neoplasia (HGPIN; n=50 cores, 25 patients), primary prostate cancer tumor (PRCA; n=141 cores, 73 patients) and metastases (METS; n=44 cores, 22 patients). To ensure high quality analysis, the pathologist chose PRCA cores containing none or <5 percent of intermixed benign prostate glands. A second “outcomes” TMA (oTMA) containing duplicate PRCA cores with associated clinical data was also stained30. Sample sizes for Gleason score (GS) groups from the oTMA are: GS 5–6 n=48, GS7 n=104, GS8–9 n=31. Sample sizes for disease free recurrence (DFS) are: DFS>5 n=136, DFS<5 n=32. Both TMAs were constructed in a single institution over the years 1998–2006, and the age range for specimens on the TMAs are from 37–86 years.
Immunohistochemistry (IHC)
Tissue sections from TMA were used in accordance to the Translational Research Initiatives in Pathology core’s standard operating procedures. Briefly, sections were cut at 5μm and stored at −80 degrees Celsius until used for staining to minimize the loss of antigenicity for AR31. Importantly, all samples compared in this experiment are on a single slide, preventing potential sectioning variability between samples. Multiplexed immunohistochemistry (IHC) was performed using the VENTANA BenchMark automated slide staining instrument (Roche Diagnostics, Basel, CHE) to detect AR (Discovery Purple chromogen), ARv7 (DAB chromogen), and smooth muscle alpha-actin (SMA, Vina Green chromogen) as previously described23. Antibodies (AR: Spring Bioscience 1:100; ARv7: Precision Antibodies 1:100, Abcam 1:200) were validated during optimization (Supplemental Figure S1). SMA was used for tissue segmentation (stroma vs. epithelium) analysis, and hematoxylin was used for cell segmentation (nuclear vs. cytoplasmic) analysis as previously described32. The protocol for this multiplexed IHC is detailed in Supplemental Figure S2. To address the potential interference between single and double stains, a comparison of intensities between single and double stains were quantified in Supplemental Figure S3.
Image Analysis
The Vectra platform (Perkin Elmer, Waltham, MA, USA) was used for automatic image acquisition. inForm™ 1.4 software, (Perkin Elmer, Waltham, MA, USA) was used for image analysis, where genitourinary pathologist (WH) trained 18% of the total images (rendering 97% accuracy) to segment nucleus from cytoplasm and epithelium from stroma. To account for cells or tissue not already identified, we established a compartment designated as “other”, which included artifact, luminal space, and non-prostate cells (i.e. nerves, red blood cells, inflammatory cells). This machine learning-based analysis created an algorithm enabling automated cell and tissue segmentation for each core sample as previously described30, 32. When this algorithm was applied to the TMAs (pTMA, oTMA), AR and ARv7 staining was objectively quantified, specifically in the nucleus of epithelial cells, through PRCA progression. To ensure tissue quality, the TMA was reviewed by a genitourinary pathologist every 10 sections confirming diagnosis of the tissue core and excluding any samples that have compromised tissue quality (i.e. tissue folding, tissue loss). Additionally, any core that contained less than 100 cells was excluded from analysis. Finally, the OD values for the 2 cores from the same patient were averaged; if one of the cores for a single patient was compromised, both cores from that patient were removed from analysis
Staining Quantification
Spectral libraries were created for each chromogen, producing a unique optical spectrum for each stain. Using these standards, the staining for each antibody was optically isolated and quantified using inForm v1.4 software. Raw segmentation data from inForm v1.4 yielded mean optical density (mean OD) values for AR and ARv7, which were calculated as the staining intensity per pixel in each core, followed by averaging the two cores from the same patient for a single mean OD per patient. AR and ARv7 “percent positivity” was quantified as the percentage of positive nuclei per core divided by the total number of nuclei in the epithelial compartment of that core. Integrating raw segmentation data from inForm v1.4 and manual thresholding of AR (mean OD threshold=0.03), ARv7 (mean OD threshold=0.155), and SMA (mean OD threshold=0.08), we calculated the proportion of each core that expressed the AR+/ARv7+ cell population. To establish the threshold for positivity, the subset of images used for software training was optically separated into individual chromogens, and visually assessed by the pathologist to determine the lower limit of positivity for select individual nuclei. This analysis of OD in individual nuclei is achieved using the “view component data” function of inForm software by hovering the cursor over an individual nucleus, which displays 1) a table of all signal intensities found at that pixel and 2) average signal intensity for each component (i.e nucleus, cytoplasm, or membrane) found. Because the optical intensity is dependent on the color of the chromogen, the thresholds for each chromogen were determined separately. Using these thresholds, we also quantified the percentage of the AR+/ARv7−, AR−/ARv7+, and AR−/ARv7− cell populations. Similarly, application of increasing positivity thresholds allowed stratification of staining intensity into AR not detected (mean OD threshold<0.02), AR low (mean OD threshold>0.02, <0.03), and AR high (mean OD threshold>0.03).
Statistical Analysis
Graphpad Prism 5.04 (Graphpad Software, La Jolla, CA, USA) was used for statistical analysis. We assessed differences among continuous variables with one-way ANOVA. Tukey’s Multiple Comparison Test was used to determine mean differences of HGPIN, PRCA, and METS compared to BPT, and difference between GS5–6, GS7, GS8–9. A Student’s T-test was used to determine differences between DFS<5 and DFS>5. Data in bar graphs and tables show mean +/− standard error of the mean. For all analyses, p< 0.05 was considered statistically significant, with * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
RESULTS
Multiplexing Immunohistochemistry to Assess Expression and Localization in situ
To assess the expression of AR and ARv7 in the same tissue samples, we co-stained a tissue microarray containing 394 human prostate samples from four stages of PRCA progression: benign prostatic tissue, high grade intraepithelial neoplasia (HGPIN), PRCA, and metastases (METS). Using a multispectral imaging system, we optically isolated chromogens to view the staining pattern for both targets (AR and ARv7) from within the multiplexed core (Figure 1A–B). Additionally, we used hematoxylin as a nuclear inclusion dye to segment the nuclear subcellular compartment (Figure 1C–D). Using this segmentation technology, we further assessed AR and ARv7 expression in prostate nuclei, where regulatory activities of these transcription factors occur. Finally, we used smooth muscle alpha actin staining (data not shown) to train the software to segment tissue types (stroma, epithelium) within each prostate core (Figure 1E). Because androgen receptor elicits effects from both the epithelial and stromal cells, this tissue segmentation allows us to assess each of these compartments independently to ensure robust and specific co-expression data with ARv7. These multispectral imaging and segmentation techniques serve as a proof of principle for the accuracy and sensitivity of the software that will be utilized to objectively assess AR and ARv7 expression and localization in human prostate samples.
FIGURE 1. AR and ARv7 in Multiplexed IHC with Cell and Tissue Segmentation.
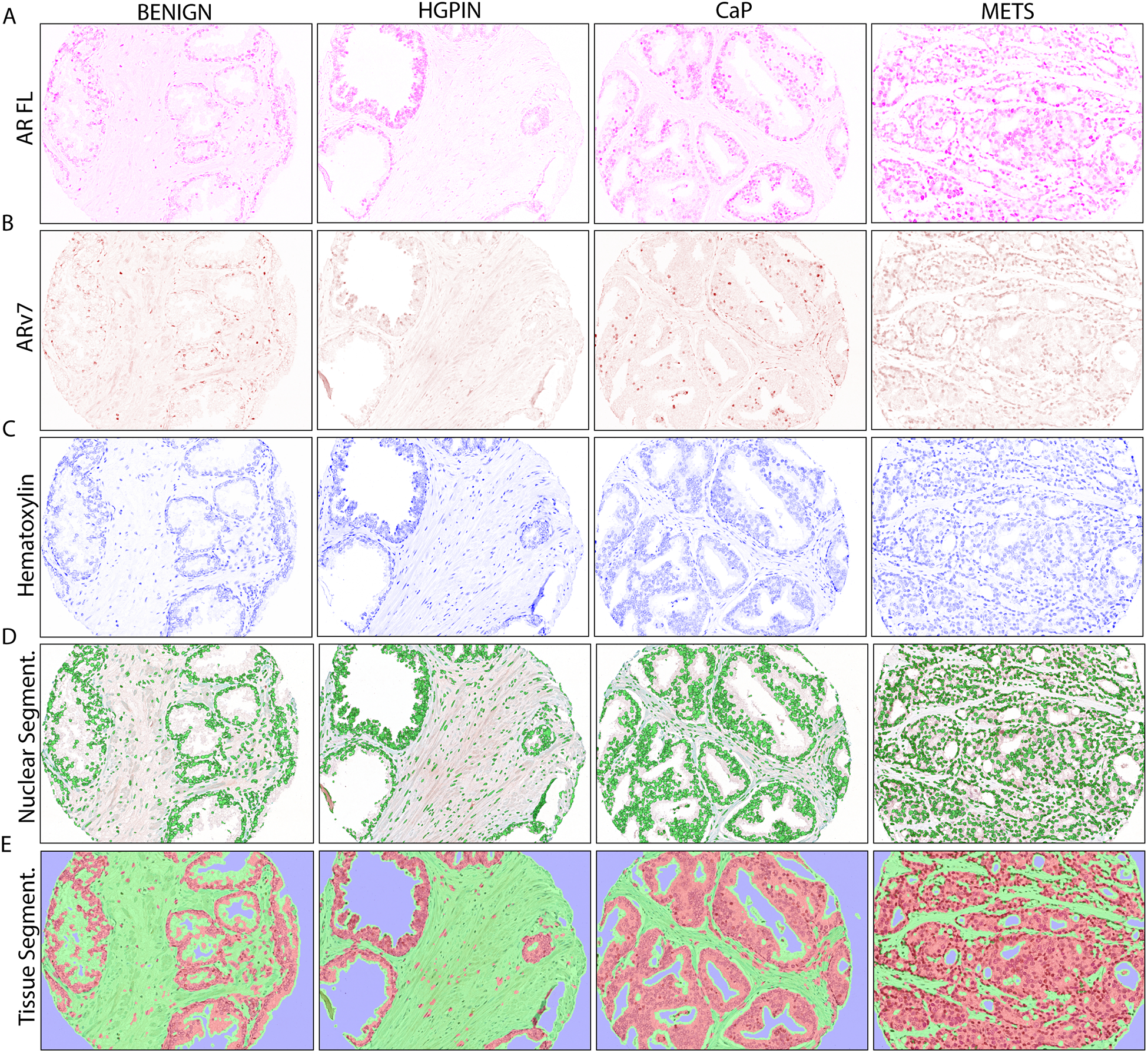
A. Optically isolated full-length AR (AR FL) staining (purple) in PRCA progression.
B. Optically isolated ARv7 staining (brown) in PRCA progression.
C. Optically isolated hematoxylin staining (blue) in PRCA progression.
D. Automated nuclear segmentation (green) in PRCA progression.
E. Automated tissue segmentation (epithelium=red, stroma=green) in PRCA progression.
BENIGN = benign prostatic tissue, n=52; HGPIN = high grade prostatic neoplasia, n=25; PRCA = prostate cancer, n=73; METS = metastasis, n=22. IHC image magnification = 20X.
Independent expression and localization of AR and ARv7 in prostate cancer progression
To quantify independent expression of AR and ARv7 in prostate cancer progression, we measured the optical density and percent positivity of AR and ARv7 positive cells within each prostate tissue core. Visual representation of the AR expression pattern in representative cores show strong nuclear staining at all stages of PRCA progression, supporting previously published data (Figure 2A). Importantly, to ensure quantification of the full-length AR alone, the AR antibody used for this study is specific for the C-terminus of the full-length AR protein, which does not detect AR variants. Quantification of full-length AR, shows nuclear AR expression significantly increased in PRCA and METS compared to benign (p<0.05) (Figure 2B). This increase was seen with two types of analysis: 1) mean optical density, which objectively measures the absolute expression of AR in each core, and 2) percent positivity, which is based on an algorithm to determine a threshold for differentiation of positivity and negativity for full-length AR expression. Notably, the trend for each type of analysis is the same; this supports the validity of the threshold used to determine percent positivity, which will be used in subsequent analysis of AR/ARv7 co-localization. Concurrent with full-length AR staining, the same tissue sections were also stained for ARv7 and quantified using the same analysis techniques. Visual representation of the ARv7 expression pattern in representative cores showed nuclear staining at all stages of PRCA progression including early stages, which has only been partially previously described (Figure 2C). When ARv7 expression was quantified, both the absolute expression and percent positivity of ARv7 is increased in METS compared to benign prostate, as expected (Figure 2D). Interestingly, there was also an increase of ARv7 in HGPIN samples compared to benign prostate, which has not been previously described (Figure 2D). Taken together, these data indicate that both AR and ARv7 exhibit expression changes through PRCA progression, where both are significantly increased in METS compared to benign prostate.
FIGURE 2. Independent AR and ARv7 Expression in PRCA Progression.
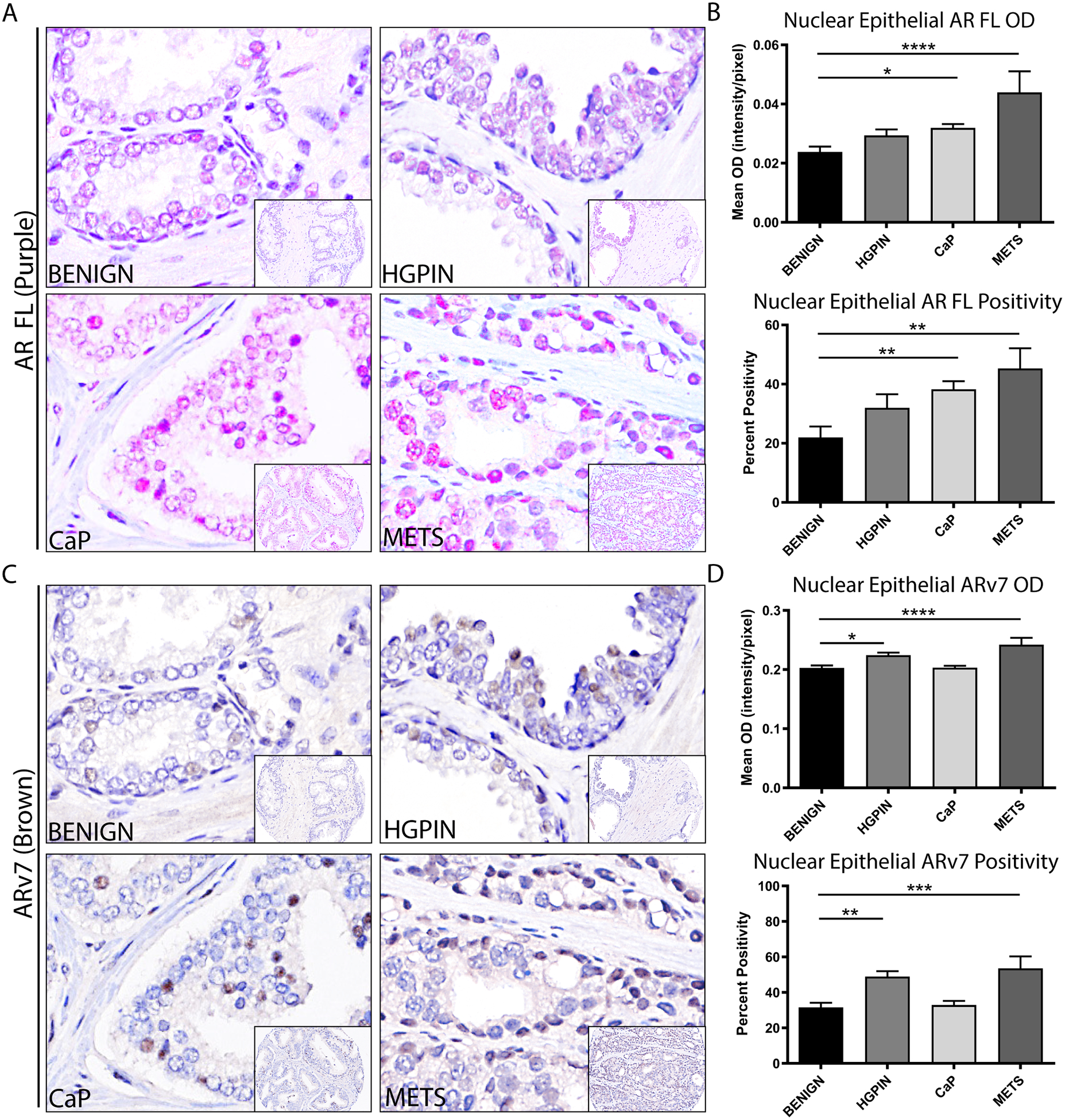
A. Representative images of full-length AR (AR FL, purple) with hematoxylin (blue), showing nuclear localization at all stages of PRCA progression.
B. Quantification of nuclear epithelial full-length AR (AR FL) in PRCA progression by mean optical density (top) and percent positivity (bottom) showed a significant increase in AR in PRCA and METS compared to BENIGN (p<0.05, p<0.0001 respectively for mean OD; p<0.01 for percent positivity).
C. Representative images of ARv7 (brown) with hematoxylin (blue), showing nuclear localization at all stages of PRCA progression.
D. Quantification of nuclear epithelial ARv7 in PRCA progression by mean optical density (top) and percent positivity (bottom) showed a significant increase in ARv7 in HGPIN and METS compared to BENIGN (p<0.05, p<0.0001 respectively for mean OD; p<0.01, p<0.001 respectively for percent positivity).
Image magnification = 100X with 20X inset.
Inter- and intra-tumoral AR Heterogeneity in Prostate Cancer
Despite the overall increase of AR and ARv7 expression in PRCA progression, cell populations that were not considered AR-positive persisted at each stage of PRCA progression, suggesting both intra- and inter-tumoral variability of AR expression. This phenomenon, known as AR heterogeneity, has recently been implicated as a predictor for clinical outcomes and therapy resistance33, 34. In this study, we objectively assessed the incidence of inter-tumoral AR heterogeneity by categorizing PRCA samples into “AR high”, “AR low”, or “AR not detected” based on increasing thresholds applied to mean optical density of each core as a whole. Visual representation of these three categories of AR expression are shown in Figure 3A. Quantification of inter-tumoral heterogeneity showed 57.35% of PRCA samples express high AR, 27.94% express low AR, and 14.71% express AR below the limit of detection (Figure 3B). The threshold used for “high AR” was the same as the threshold for percent positivity in Figure 2B, resulting in consistency of positivity between the two analyses. Interestingly, within the subset of PRCA cores that are not considered AR positive in Figure 2, there are two distinct expression patterns that we observed: 1) low AR, representing cores that express AR below the threshold considered to be positive, and 2) AR not detected, where AR protein was not identified. In addition to inter-tumoral heterogeneity, we also observed intra-tumoral heterogeneity of AR expression within individual nuclei of select PRCA cores using the “view component data” function on inForm. Here we saw varying levels of AR expression within a single core (Figure 3C). Taken together, these data show that with objective quantification of AR expression, we can categorize AR expression into three distinct levels providing evidence for AR heterogeneity in PRCA.
FIGURE 3. Inter- and Intra-tumoral Heterogeneity of AR in PRCA Progression.
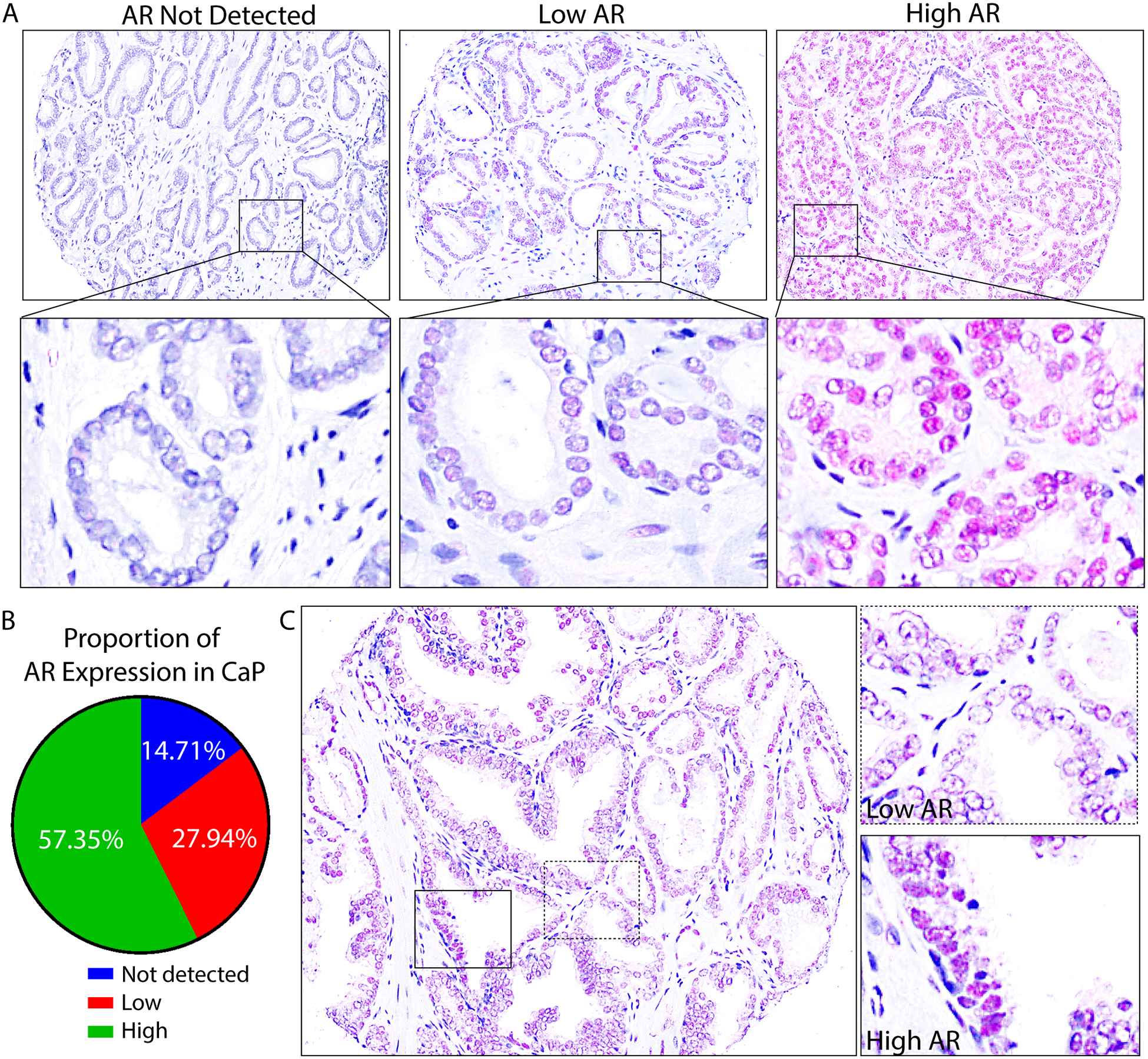
A. Composite images of full-length AR in nuclei of epithelial cells (purple) in PRCA cores showed inter-tumoral heterogeneity of AR: AR not detected, low AR, and high AR expressing cores.
B. The percentage of cores expressing high AR, low AR, and core where AR was not detected were quantified among PRCA cores in the pTMA (n=73) and represented in a pie chart. 57.35% express high levels of AR, 27.94% expressed low AR, and 14.71% did not have detectable AR.
C. An example PRCA core shows intra-tumoral AR heterogeneity, where AR expression is shown in purple. The full core (20X) is shown on the left, and high magnification (100X) images are shown on the right. AR expression ranges from high (solid box) to low (dashed box).
AR and ARv7 Co-expression in Prostate Cancer Progression
AR and ARv7 have been shown to interact as a heterodimer in PRCA, yet the incidence and stage specificity of these receptors’ co-expression has not been determined or quantified on a per cell basis. To assess the co-expression of AR and ARv7 in PRCA progression, we used a machine-learning software (inForm) to quantify the percent of each core that expressed 1) AR, but not ARv7 (AR+/ARv7−), 2) ARv7, but not AR (AR−/ARv7+), 3) both AR and ARv7 (AR+/ARv7+), or 4) neither AR nor ARv7 (AR−/ARv7−). Representative images show each of these cell populations pseudo-colored (red: AR+/ARv7−, green: AR/ARv7+, yellow: AR+/ARv7+, blue: AR−/ARv7−) in PRCA progression tissue cores (Figure 4A). Here, we saw a significant increase of AR+/ARv7− in PRCA and AR−/ARv7+ in METS compared to benign (p<0.01 and p<0.001, respectively) (Figure 4B–C). The AR+/ARv7+ population significantly increased in METS compared to benign (p<0.05), and was trending toward a significant increase in HGPIN compared to benign (p=0.07) (Figure 4D). Interestingly, the population that lacked both receptors, AR−/ARv7−, was also increased in PRCA compared to benign (p<0.05) (Figure 4E). Taken together, these data provide evidence of four distinct PRCA cell populations in terms of AR expression: double positive, AR only, ARv7 only, and double negative. Additionally, the incidences of AR/ARv7 co-expressing and ARv7 only cells were increased in METS, suggesting these populations could contribute to metastatic cell growth. Likewise, the incidence of AR only and double negative cells increased in PRCA, suggesting these populations may play a role in the progression of PRCA.
FIGURE 4. AR and ARv7 Co-expression in PRCA Progression.
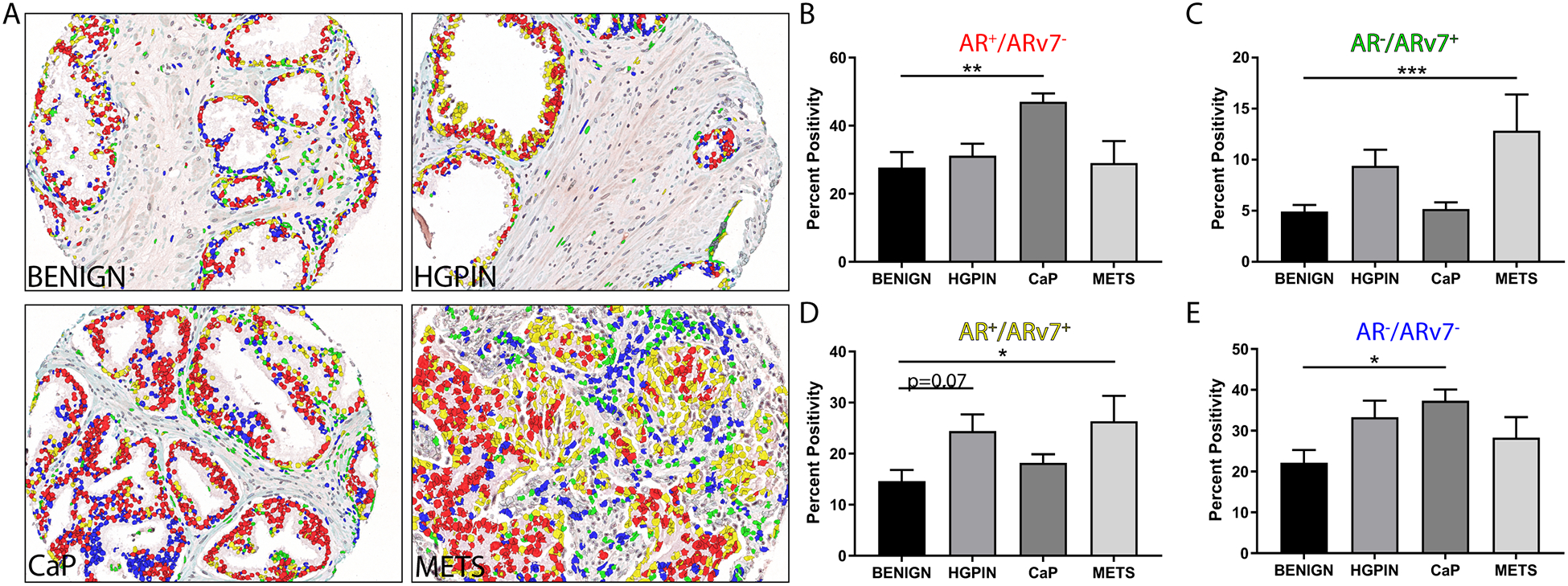
A. Prostate composite images show AR+/ARv7− nuclei (red), AR−/ARv7+ nuclei (green), AR+/ARv7+ nuclei (yellow), and AR−/ARv7− nuclei (blue) at all stages of PRCA progression.
B. Quantification of the AR+/ARv7− population (red) shows a significant increase in PRCA compared to BENIGN (p<0.01).
C. Quantification of the AR−/ARv7+ population (green) shows a significant increase in METS compared to BENIGN (p<0.001).
D. Quantification of the AR+/ARv7+ population (yellow) shows a significant increase in METS compared to BENIGN (p<0.05), and a slight increase in HGPIN compared to BENIGN (p=0.07).
E. Prostate composite images show AR−/ARv7− nuclei (blue) shows a significant increase in PRCA compared to BENIGN (p<0.05).
AR and ARv7 co-expression predicts advanced Gleason scores and disease recurrence
To assess clinical implications of AR/ARv7 co-expression, we co-stained a TMA containing samples with associated patient data for clinical outcomes (oTMA). Using the same multispectral imaging and machine learning software that was used to analyze the pTMA, we quantified AR and ARv7 expression in the oTMA. Two endpoints assessed showed significant changes in AR/ARv7 expression that may be clinically relevant: Gleason score (GS) and disease-free survival time (DFS). Gleason grading is a histology-based system for PRCA tumors where higher Gleason score (GS) represents more advanced tumors (i.e. decreased glandular differentiation, increased infiltration)35. Representative images show cores from tumors with Gleason score 5–6 (GS5–6), 7 (GS7), and 8–9 (GS8–9) at the time of tissue collection (Figure 5A). Similar to the co-expression analysis for the pTMA, these images were pseudo-colored to represent cell populations that express AR+/ARv7− (red), AR−/ARv7+ (green), AR+/ARv7+ (yellow), and AR−/ARv7− (blue) (Figure 5A). We observed no differences in AR+/ARv7− and AR−/ARv7+ populations in GS8–9 tumors compared to lower GS tumors (Figure 5B). However, AR+/ARv7+ double positive cells significantly decreased in GS8–9 relative to GS7 (p<0.05), yet they were not different compared to GS5–6 (p=0.11) (Figure 5B). Strikingly, the double negative, AR−/ARv7−, population was significantly increased in GS8–9 compared to both GS5–6 and GS7 subgroups (p<0.01) (Figure 5B). Recently, an updated version of this classification system, Grade Grouping, has been established based on overall survival36; with analysis using this grading system, we observed similar results to analysis with GS (Supplemental Table 1). Next, we assessed disease-free survival time, which was measured as the number of years post-collection before the cancer was considered recurrent. This endpoint is of particular importance because it represents the staining pattern in tissues that will become recurrent, rather than biopsies from recurrent tissue, suggesting any expression changes may serve as a predictor of recurrence. Representative images show example cores from recurrence that occurred within 5 years (DFS<5) and recurrence that occurred more than 5 years (DFS>5) after tissue collection, with cell populations pseudo-colored as described above (Figure 5C). Here, we saw no change in expression of AR or ARv7 alone between DFS<5 and DFS>5 years (Figure 5D). Interestingly, we saw a significant decrease in the AR+/ARv7+ population (p<0.01) and a significant increase in the AR−/ARv7− population (p<0.05) in DFS<5 compared to DFS>5 (Figure 5D). These data support the concept that PRCAs that lack protein expression of both AR and ARv7 become recurrent earlier than PRCAs that co-express AR and ARv7. Taken together, these clinical outcomes data suggest that assessing AR or its variants alone may not have prognostic value; however, assessing the receptor co-expression, or lack thereof, may be 1) an indicator of advanced cancer, corroborated by higher GS, and 2) a predictor for cancers that will become recurrent.
FIGURE 5. AR and ARv7 Co-expression and Clinical Outcomes.
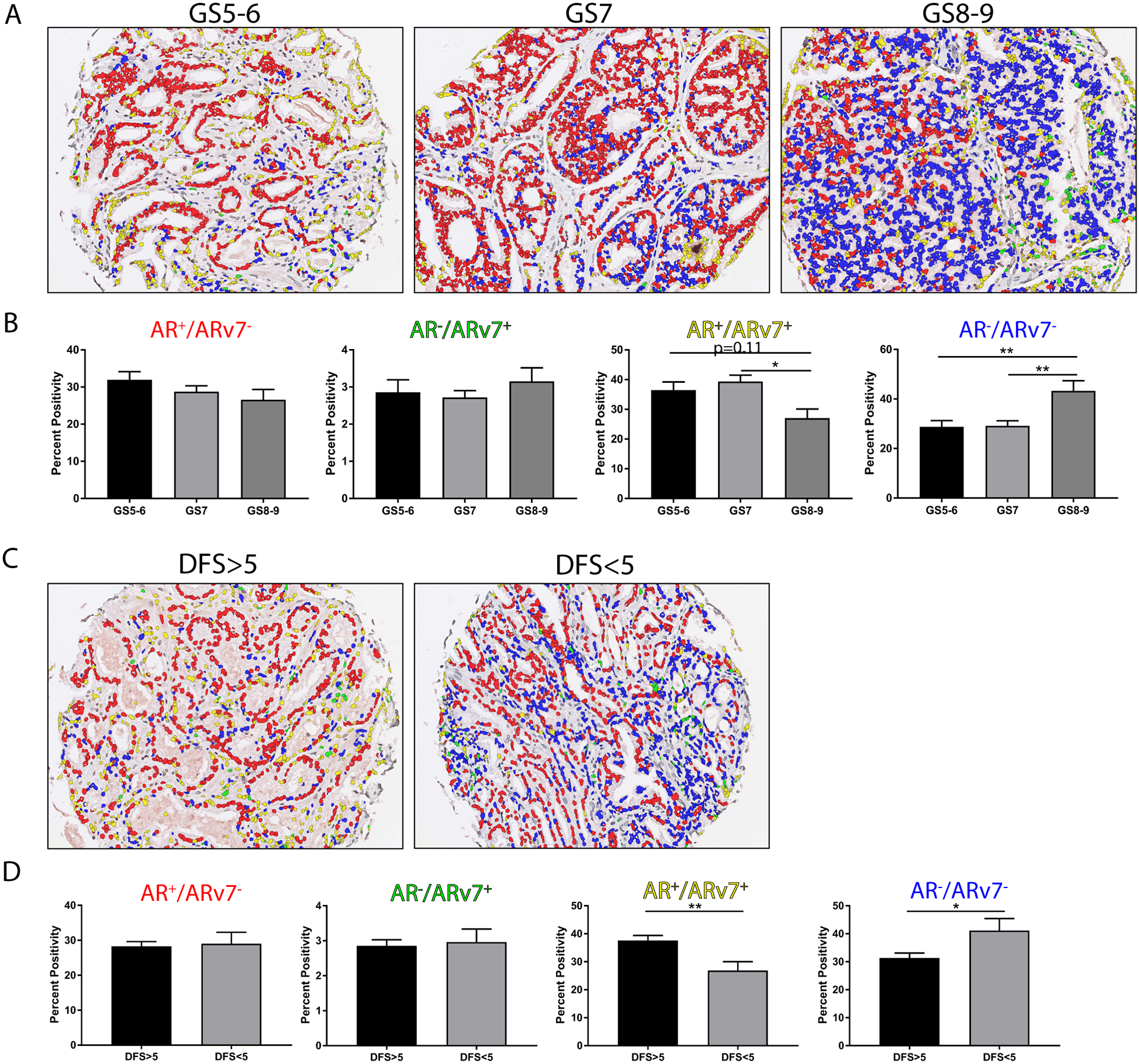
A. Prostate composite images show AR+/ARv7− nuclei (red), AR−/ARv7+ nuclei (green), AR+/ARv7+ nuclei (yellow), and AR−/ARv7− nuclei (blue) separated by Gleason score (GS): GS5–6 n=48, GS7 n=104, GS8–9 n=31).
B. Quantification of the percent positivity of the AR+/ARv7− (red), AR−/ARv7+ (green), AR+/ARv7+ (yellow), AR−/ARv7− (blue) populations show no significant change with in the AR+/ARv7− (red) and AR/ARv7+ (green) populations. The AR+/ARv7+ (yellow) population was significantly decreased in GS8–9 compared to GS7 (p<0.05), and trending toward significant decrease compared to GS5–6 (p=0.11). The AR−/ARv7− (blue) population was significantly increased in GS8–9 compared to GS7 and GS5–6 (p<0.01 for both).
C. Prostate composite images show AR+/ARv7− nuclei (red), AR−/ARv7+ nuclei (green), AR+/ARv7+ nuclei (yellow), and AR−/ARv7− nuclei (blue) separated by disease free survival greater than 5 years (DFS>5 n=136) or less than 5 years (DFS<5 n=32).
D. Quantification of the percent positivity of the AR+/ARv7− (red), AR−/ARv7+ (green), AR+/ARv7+ (yellow), AR−/ARv7− (blue) populations show no significant change with in the AR+/ARv7− (red) and AR/ARv7+ (green) populations. The AR+/ARv7+ (yellow) population was significantly decreased in DFS<5 compared to DFS>5 (p<0.01). The AR−/ARv7− (blue) population was significantly increased in DFS<5 compared to DFS>5 (p<0.05).
DISCUSSION
Prostate cancer is known to be driven by androgen signaling, yet the incidence of AR expression in PRCA progression and disease recurrence is still controversial. With the discovery of AR splice variants, there has been a growing interest in expression patterns of full-length AR and its variants in disease progression, and how these receptors, independently and together, contribute to adverse clinicopathologic outcomes. To objectively quantify AR and ARv7 in human PRCA samples, we used multiplexed IHC to co-stain with AR and ARv7 and analyzed expression changes with a machine-learning based software; understanding the prevalence and clinicopathologic outcomes associated with changes in these receptors’ expression may improve diagnosis and ultimately treatment for men with PRCA.
This study is strengthened by the use of techniques and reagents that have only recently become available. Technical advances include the use of multiplexed IHC, multispectral imaging, and machine-learning software to objectively quantify expression of multiple proteins in the same sample (Figure 1). Co-expression studies using immunostaining on fixed tissue has historically been difficult; however, this study was able to overcome these technical challenges by using an automated multiplexing technique that supports data reproducibility30, 32. Moreover, the analysis software used in this study allows the segmentation of tissue compartments into epithelium and stroma, and the segmentation of the cellular compartments into nuclear and cytoplasmic (Figure 1)23. This is significant because the regulatory effects of AR and ARv7 occur in the nucleus and PRCA is primarily composed of epithelial cells37. Using this analysis platform, we can focus on transcriptionally active epithelial AR at the exclusion of stromal and cytoplasmic AR. Additionally, advancement of reagents include ARv7-specific antibodies, and AR C-terminal antibodies that specifically detect the full-length protein but not AR variants. All the antibodies used in this study are commercially available and validated by the experiments herein, the manufacturers, and in peer-reviewed literature18, 20, 38. With these advancements in techniques and reagents, it is possible to 1) objectively quantify receptor expression trends in PRCA progression, potentially mitigating interexperimental variability and increase reproducibility, 2) specifically assess expression in the tissue/cell compartment of interest to increase specificity, and 3) distinguish full-length AR expression from AR variant expression to accurately quantify each individually.
Using these techniques and reagents, we founds several interesting expression changes in AR and ARv7 in PRCA progression. First, we were able to repeat the previously published expression trends for AR and ARv7, where both were increased in METS compared to benign (Figure 2)22, 23, 28. This data reproduction serves as a validation for the antibodies chosen in this study, and for the efficacy of the techniques. Second, we noted a persistence of cells that expressed low or undetectable AR at each stage of PRCA progression, which has previously been shown23 (Figure 3). While it is generally accepted that there are cell populations within the normal prostate that do not express AR37, this AR-low cell population in the later stages of PRCA progression may be of interest in terms of anti-androgen therapy resistance39. This concept of AR heterogeneity has previously been associated with poor outcomes in patient survival, and this study provides evidence that these cell populations are present in human PRCA33, 34. Finally, this study showed an increase of ARv7 in metastatic PRCA, as expected28. Interestingly, we detected ARv7 positivity at every stage of PRCA progression, in contrast to others27. This inconsistency could be due to differing reagents and/or analysis techniques. Though low in abundance at earlier stages of PRCA progression, these ARv7-positive cells may represent a cell population that contributes to disease recurrence. Taken together, perhaps the cell populations identified in this study that lack AR expression or express ARv7 at earlier stages of PRCA progression are intrinsically resistant to AR-targeted therapeutics, and further investigation of these populations could provide insight into therapeutically targetable proteins or pathways that could reduce disease recurrence.
Another analysis technique that became available with the use of machine-learning based software is quantification of AR and ARv7 co-expression (Figure 4). This expression pattern is of interest due to the discovery of AR/ARv7 interaction as heterodimers19. Importantly, steroid hormone receptor heterodimers have been shown to transcriptionally activate a distinct subset of downstream targets, which may affect the efficacy of certain therapies20, 40, 41. In this study, we showed AR/ARv7 double positivity increasing in metastatic PRCA (Figure 4), suggesting there may be heterodimerization in advanced stages of PRCA progression - this interaction could be a topic of interest in future studies. Additionally, this analysis technique allowed quantification of the cell population that lacked both AR and ARv7. This double negative population could be of importance due to the implications of antiandrogen therapy resistance mentioned previously. Our data showed an increase in this population in PRCA compared to benign, perhaps suggesting a clonal expansion of this population in PRCA progression (Figure 4). To our knowledge, this is the first study to quantify AR and ARv7 simultaneously in the same cell through PRCA progression in this manner. Results from this study may offer a better understanding of the role of AR and ARv7 in PRCA progression, and how co-expression of these receptors may alter the efficacy of AR-targeted therapeutics.
To specifically address the role AR/ARv7 co-expression in the context of patient outcomes, we quantified AR and ARv7 expression in PRCA samples with associated patient data (Figure 5). These results suggest that analysis of AR and ARv7 expression independently is not sufficient to predict Gleason grade or time to recurrence (i.e. disease-free survival). This contradicts previous studies, where ARv7 positivity predicts reduced PSA-progression free survival17; this inconsistency could be due to different measures of 1) recurrence (PSA-progression free survival vs. disease-free survival) or 2) ARv7 positivity (mRNA vs. protein). Interestingly, in this study, receptor co-expression exhibited significant changes with adverse clinical outcomes. The AR+/ARv7+ cell population is significantly decreased with increased Gleason grade, while the AR−/ARv7− population is significantly increased (Figure 5). This suggests that more aggressive tumors express lower levels of AR and ARv7 than tumors that are considered less aggressive, perhaps supporting the idea of AR heterogeneity and AR-negative clonal expansion as tumors progress42. Additionally, the AR+/ARv7+ cell population is significantly decreased in tumors that become recurrent in under 5 years, while the AR−/ARv7− population is significantly increased (Figure 5). This suggests the level of AR expression is an indicator of time to recurrence, perhaps due to the widespread use of therapies that directly target AR (i.e. bicalutamide, enzalutamide, apalutamide)43–45. It is conceivable that under ADT conditions, the AR−/ARv7− population would not respond to therapy due to the lack of targetable AR protein. Therefore, it may be important to identify therapeutically targetable agents in this AR−/ARv7− population to prevent recurrent tumor growth.
The advantages of machine-learning based analysis of immunogenic co-staining are multiple; however, there are some potential weaknesses to this emerging technique. First, the efficacy of the automated tissue and cell segmentation relies on an algorithm that is created from a subset of the cores. While it is unlikely that this subset of cores use for software “training” is not a representative sample set, it is possible that the algorithm is not accurate in every core in the TMAs. Second, the associations between stage, Gleason score, recurrence and protein expression are correlative. Therefore, significant changes in overall expression may not be functionally relevant to disease progression or recurrence, and further experimentation would be needed to investigate these functional implications. Third, AR positivity in this study was assessed using thresholds that were determined based off optical density in individual nuclei; because there is no consensus for the expression levels of AR necessary for biological activity, these thresholds are subjective and may not represent AR activity. Additionally, the staining patterns may have been affected by the fixation, sectioning, or staining protocols used for this study (i.e antigenicity of epitopes), which should be addressed in future studies. Fourth, normalization of epithelial staining intensity for these types of studies is frequently compared to the stromal intensity for the protein of interest. However, in the case of AR, it is a well known phenomenon that stromal AR expression is decreased in later stages of PRCA progression and in recurrence46–48. These trends confound this type of normalization due to the likely exclusion of high grade or recurrent PRCAs and metastases because of a reduction in stromal AR, and were not applied in this study. Finally, the TMAs used in this study contain duplicate cores for each patient, resulting in an overall sample sizes that range from 22 to 136 patients per group; these groups are relatively small, and may not represent the true PRCA patient population or cause a bias in the data for the smaller groups. Despite these potential drawbacks, an automated, objective approach remains the preferred approach for association studies of this nature.
While a multitude of previous studies have assessed the expression and localization of AR and AR variants in PRCA, the results are inconsistent due to technical difficulties and subjective analysis. This study is feasible due to 1) the development of validated, commercially-available AR full-length and ARv7-specific antibodies, 2) the use of an automated IHC platform to allow multiplexing and prevent protocol variability, and 3) objective quantification with machine-learning software to reduce inter-observer scoring variability and hotspot analysis. Here, we show AR and ARv7 expression changes, both independently and together, through PRCA progression with implications in clinicopathologic outcomes. A more complete understanding of the role of these receptors in PRCA may provide insight into therapeutic techniques to improve patient survival.
Supplementary Material
SUPPLEMENTAL FIGURE S3. Staining Intensity Quantification Between Single and Dual Stains A. A comparison between the staining intensities of AR alone (single stain) and AR co-stained with ARv7 (double stain) on human prostate tissue shows there is no significant difference in AR staining intensity between the two type of staining (p=0.21). The staining intensity, measured in arbitrary units (AU), represents the average intensity of all nuclei within a 20x image. The data for single vs. double staining was plotted in a box and whisker plot (n=3 for both). Significance was determined using a studenťs T-test.
B. A comparison between the staining intensities of ARv7 alone (single stain) and ARv7 co-stained with full length AR (double stain) on human prostate tissue shows there is no significant difference in ARv7 staining intensity between the two type of staining (p=0.68). The staining intensity, measured in arbitrary units (AU), represents the average intensity of all nuclei within a 20x image. The data for single vs. double staining was plotted in a box and whisker plot (n=3 for both). Significance was determined using a studenťs T-test.
SUPPLEMENTAL FIGURE S2. Multiplexed IHC Staining Protocol A protocol for multiplexed staining of the TMAs including dilution buffers, incubation times, antigen retrieval methods, and order of antibody application shows details of the multiplexed staining process for reference.
SUPPLEMENTAL FIGURE S1. AR and ARv7 Antibody Validation A. Representative images of two ARv7 specific antibodies (Abcam and Precision) and full-length AR C-terminal (Spring Bioscience) staining in positive and negative control xenografts. The PRCA cell line CWR22RV1 expresses high levels of both full-length AR and ARv7. A “no primary antibody” (“primary (−)”) control was used for each antibody to show a lack of non-specific staining, while the xenografts stained with primary antibody (“primary (+)”) show nuclear positivity for each of the antibodies, as expected. The C42 cell line expresses full-length AR, but lacks ARv7 expression. When stained with each primary antibody, C42 xenografts showed positivity for full-length AR, but no positivity for either ARv7 specific antibody.
B. Representative images of human prostate samples at two stages (benign and PRCA) with both ARv7 antibodies shows little/no positivity in benign, but strong nuclear positivity for both in PRCA samples. Staining with the full-length C-terminal AR antibody shows strong nuclear positivity in both benign and PRCA samples.
ACKNOWLEDGMENTS
The authors thank the University of Wisconsin TRIP laboratory and Ms. Sally Drew for their testing and standardization of AR antibodies, and the UW-Madison Carbone Cancer Center for their financial support of core services P30 CA014520 (UWCCC). We also acknowledge the help Glen Leversen for bio-statistical assistance. Financial support was provided by National Institutes of Health grants U54 DK104310 (WAR) and R01 ES001332 (WAR). JEV is a trainee in the Cancer Biology Graduate Program at the University of Wisconsin-Madison and was funded by T32 CA009135.
Footnotes
DISCLOSURE STATEMENT: The authors have nothing to disclose.
REFERENCES
- 1.Huggins C, Hodges CV. Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. The Journal of urology. 2002;167(2 Part 2):948–951. [PubMed] [Google Scholar]
- 2.Gelmann EP. Molecular biology of the androgen receptor. Journal of Clinical Oncology. 2002;20(13):3001–3015. [DOI] [PubMed] [Google Scholar]
- 3.Sharifi N, Gulley JL, Dahut WL. An update on androgen deprivation therapy for prostate cancer. Endocrine-related cancer. 2010;17(4):R305–R315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirby M, Hirst C, Crawford E. Characterising the castration-resistant prostate cancer population: a systematic review. International journal of clinical practice. 2011;65(11):1180–1192. [DOI] [PubMed] [Google Scholar]
- 5.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32(49):5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visakorpi T, Hyytinen E, Koivisto P, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nature genetics. 1995;9(4):401. [DOI] [PubMed] [Google Scholar]
- 7.Taplin M-E, Bubley GJ, Ko Y-J, et al. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer research. 1999;59(11):2511–2515. [PubMed] [Google Scholar]
- 8.Culig Z, Hobisch A, Cronauer MV, et al. Mutant androgen receptor detected in an advanced-stage prostatic carcinoma is activated by adrenal androgens and progesterone. Molecular endocrinology. 1993;7(12):1541–1550. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X-Y, Malloy PJ, Krishnan AV, et al. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nature medicine. 2000;6(6):703. [DOI] [PubMed] [Google Scholar]
- 10.Varkaris A, Corn PG, Gaur S, Dayyani F, Logothetis CJ, Gallick GE. The role of HGF/c-Met signaling in prostate cancer progression and c-Met inhibitors in clinical trials. Expert opinion on investigational drugs. 2011;20(12):1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karan D, Kelly DL, Rizzino A, Lin M-F, Batra SK. Expression profile of differentially-regulated genes during progression of androgen-independent growth in human prostate cancer cells. Carcinogenesis. 2002;23(6):967–976. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama T, Watanabe M, Suzuki H, et al. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Laboratory investigation. 2000;80(12):1789. [DOI] [PubMed] [Google Scholar]
- 13.Hoang DT, Iczkowski KA, Kilari D, See W, Nevalainen MT. Androgen receptor-dependent and-independent mechanisms driving prostate cancer progression: opportunities for therapeutic targeting from multiple angles. Oncotarget. 2017;8(2):3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bluemn EG, Coleman IM, Lucas JM, et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell. October 9 2017;32(4):474–489 e476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocrine-related cancer. 2011;18(5):R183–R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun S, Sprenger CC, Vessella RL, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. The Journal of clinical investigation. 2010;120(8):2715–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. September 11 2014;371(11):1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer research. 2009;69(1):16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu D, Zhan Y, Qi Y, et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res. September 1 2015;75(17):3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cato L, de Tribolet-Hardy J, Lee I, et al. ARv7 Represses Tumor-Suppressor Genes in Castration-Resistant Prostate Cancer. Cancer cell. 2019;35(3):401–413. e406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchanan G, Ricciardelli C, Harris JM, et al. Control of Androgen Receptor Signaling in Prostate Cancer by the Cochaperone Small Glutamine-Rich Tetratricopeptide Repeat Containing Protein α. Cancer research. 2007;67(20):10087–10096. [DOI] [PubMed] [Google Scholar]
- 22.Gregory CW, He B, Johnson RT, et al. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer research. 2001;61(11):4315–4319. [PubMed] [Google Scholar]
- 23.Sehgal PD, Bauman TM, Nicholson TM, et al. Tissue-specific quantification and localization of androgen and estrogen receptors in prostate Cancer. Human pathology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Wheeler T, Dai H, Frolov A, Thompson T, Ayala G. High level of androgen receptor is associated with aggressive clinicopathologic features and decreased biochemical recurrence-free survival in prostate: cancer patients treated with radical prostatectomy. The American journal of surgical pathology. 2004;28(7):928–934. [DOI] [PubMed] [Google Scholar]
- 25.Mohler JL, Chen Y, Hamil K, et al. Androgen and glucocorticoid receptors in the stroma and epithelium of prostatic hyperplasia and carcinoma. Clinical Cancer Research. 1996;2(5):889–895. [PubMed] [Google Scholar]
- 26.Chodak GW, Kranc DM, Puy LA, Takeda H, Johnson K, Chang C. Nuclear localization of androgen receptor in heterogeneous samples of normal, hyperplastic and neoplastic human prostate. The Journal of urology. 1992;147(3 Part 2):798–803. [DOI] [PubMed] [Google Scholar]
- 27.Sharp A, Coleman I, Yuan W, et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. The Journal of clinical investigation. 2018;129(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Morrissey C, Sun S, et al. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PloS one. 2011;6(11):e27970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vellky JE, Ricke EA, Huang W, Ricke WA. Expression and Localization of DDX3 in Prostate Cancer Progression and Metastasis. The American Journal of Pathology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Hennrick K, Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Human pathology. 2013;44(1):29–38. [DOI] [PubMed] [Google Scholar]
- 31.Andeen NK, Bowman R, Baullinger T, Brooks JM, Tretiakova MS. Epitope Preservation Methods for Tissue MicroarraysLongitudinal Prospective Study. American journal of clinical pathology. 2017;148(5):380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauman TM, Ricke EA, Drew SA, Huang W, Ricke WA. Quantitation of protein expression and co-localization using multiplexed immuno-histochemical staining and multispectral imaging. JoVE (Journal of Visualized Experiments). 2016(110):e53837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeda H, Akakura K, Masai M, Akimoto S, Yatani R, Shimazaki J. Androgen receptor content of prostate carcinoma cells estimated by immunohistochemistry is related to prognosis of patients with stage D2 prostate carcinoma. Cancer: Interdisciplinary International Journal of the American Cancer Society. 1996;77(5):934–940. [PubMed] [Google Scholar]
- 34.De Winter JAR, Trapman J, Brinkmann AO, et al. Androgen receptor heterogeneity in human prostatic carcinomas visualized by immunohistochemistry. The Journal of pathology. 1990;160(4):329–332. [DOI] [PubMed] [Google Scholar]
- 35.Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. The Journal of urology. 1974;111(1):58–64. [DOI] [PubMed] [Google Scholar]
- 36.Erickson A, Sandeman K, Lahdensuo K, et al. New prostate cancer grade grouping system predicts survival after radical prostatectomy. Human pathology. 2018;75:159–166. [DOI] [PubMed] [Google Scholar]
- 37.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25(2):276–308. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Wang Z, Xiao W, et al. Androgen-receptor splice variant-7-positive prostate cancer: a novel molecular subtype with markedly worse androgen-deprivation therapy outcomes in newly diagnosed patients. Modern Pathology. 2018;31(1):198. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Deng Q, Chao HP, et al. Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses. Nat Commun. September 6 2018;9(1):3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowley SM, Hoare S, Mosselman S, Parker MG. Estrogen receptors α and β form heterodimers on DNA. Journal of Biological Chemistry. 1997;272(32):19858–19862. [DOI] [PubMed] [Google Scholar]
- 41.Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, Spelsberg TC. Estrogen receptor α and β heterodimers exert unique effects on estrogen-and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Molecular Endocrinology. 2005;19(6):1555–1568. [DOI] [PubMed] [Google Scholar]
- 42.Skvortsov S, Skvortsova II, Tang DG, Dubrovska A. Concise review: Prostate cancer stem cells: Current understanding. Stem Cells. 2018;36(10):1457–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masiello D, Cheng S, Bubley GJ, Lu ML, Balk SP. Bicalutamide functions as an androgen receptor antagonist by assembly of a transcriptionally inactive receptor. Journal of Biological Chemistry. 2002;277(29):26321–26326. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman-Censits J, Kelly WK. Enzalutamide: a novel antiandrogen for patients with castrate-resistant prostate cancer. Clinical Cancer Research. 2013;19(6):1335–1339. [DOI] [PubMed] [Google Scholar]
- 45.Clegg NJ, Wongvipat J, Joseph JD, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer research. 2012;72(6):1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henshall SM, Quinn DI, Lee CS, et al. Altered expression of androgen receptor in the malignant epithelium and adjacent stroma is associated with early relapse in prostate cancer. Cancer research. 2001;61(2):423–427. [PubMed] [Google Scholar]
- 47.Wikström P, Marusic J, Stattin P, Bergh A. Low stroma androgen receptor level in normal and tumor prostate tissue is related to poor outcome in prostate cancer patients. The Prostate. 2009;69(8):799–809. [DOI] [PubMed] [Google Scholar]
- 48.Ricciardelli C, Choong CS, Buchanan G, et al. Androgen receptor levels in prostate cancer epithelial and peritumoral stromal cells identify non-organ confined disease. The Prostate. 2005;63(1):19–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL FIGURE S3. Staining Intensity Quantification Between Single and Dual Stains A. A comparison between the staining intensities of AR alone (single stain) and AR co-stained with ARv7 (double stain) on human prostate tissue shows there is no significant difference in AR staining intensity between the two type of staining (p=0.21). The staining intensity, measured in arbitrary units (AU), represents the average intensity of all nuclei within a 20x image. The data for single vs. double staining was plotted in a box and whisker plot (n=3 for both). Significance was determined using a studenťs T-test.
B. A comparison between the staining intensities of ARv7 alone (single stain) and ARv7 co-stained with full length AR (double stain) on human prostate tissue shows there is no significant difference in ARv7 staining intensity between the two type of staining (p=0.68). The staining intensity, measured in arbitrary units (AU), represents the average intensity of all nuclei within a 20x image. The data for single vs. double staining was plotted in a box and whisker plot (n=3 for both). Significance was determined using a studenťs T-test.
SUPPLEMENTAL FIGURE S2. Multiplexed IHC Staining Protocol A protocol for multiplexed staining of the TMAs including dilution buffers, incubation times, antigen retrieval methods, and order of antibody application shows details of the multiplexed staining process for reference.
SUPPLEMENTAL FIGURE S1. AR and ARv7 Antibody Validation A. Representative images of two ARv7 specific antibodies (Abcam and Precision) and full-length AR C-terminal (Spring Bioscience) staining in positive and negative control xenografts. The PRCA cell line CWR22RV1 expresses high levels of both full-length AR and ARv7. A “no primary antibody” (“primary (−)”) control was used for each antibody to show a lack of non-specific staining, while the xenografts stained with primary antibody (“primary (+)”) show nuclear positivity for each of the antibodies, as expected. The C42 cell line expresses full-length AR, but lacks ARv7 expression. When stained with each primary antibody, C42 xenografts showed positivity for full-length AR, but no positivity for either ARv7 specific antibody.
B. Representative images of human prostate samples at two stages (benign and PRCA) with both ARv7 antibodies shows little/no positivity in benign, but strong nuclear positivity for both in PRCA samples. Staining with the full-length C-terminal AR antibody shows strong nuclear positivity in both benign and PRCA samples.


