Figure S1.
Related to Figure 1
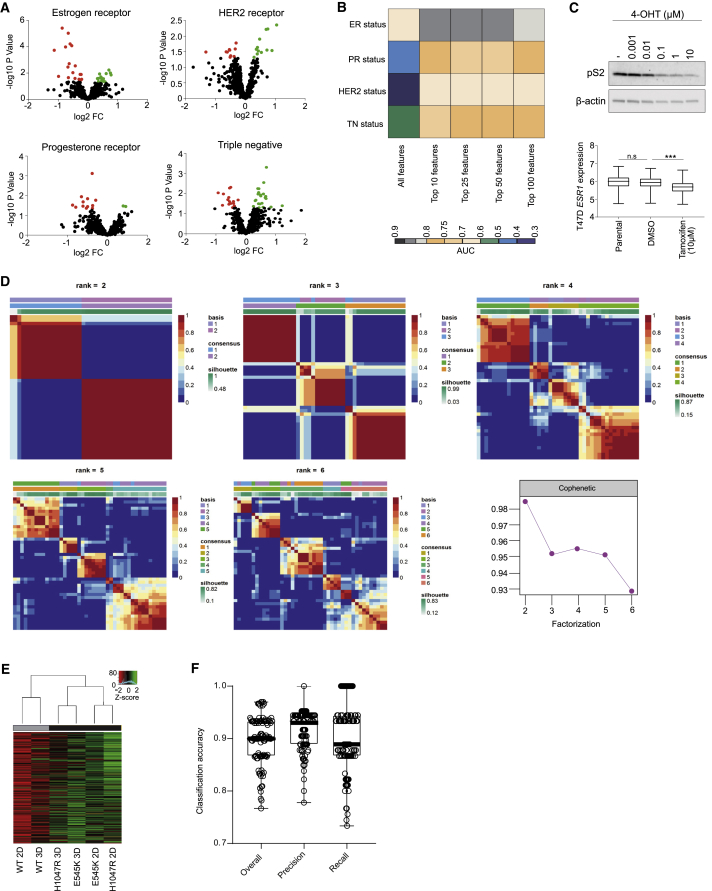
(A) Volcano plots of significantly altered phospholipids between receptor positive and negative cell lines. Black dots: not significantly altered; Red dots: significantly upregulated; Green dots: significantly downregulated phospholipids. (B) Area under the curve (AUC) classification accuracies for estrogen (ER), progesterone (PR), HER2 receptor and triple negative status of 30 primary and PDX breast tumors (median intensity of n = 3 separate sections per tumor) following feature selection for phospholipids in the m/z range 600-900 and leave-one-out cross validation. (C) Immunoblot analysis of estrogen inducible protein pS2 (top) and prediction of ESR1 expression (bottom) in ER+ve T47D cells following treatment with 0.1% DMSO or indicated concentrations of 4-OHT for 72 hours using REIMS. (D) NMF consensus maps summarizing the clustering of cell lines used in Figure 1D. The color map represents the correlation between cell lines in the same cluster when samples are divided into 2-6 groups. The highest cophenetic score was obtained for two clusters. (E) REIMS analysis of MCF10A PIK3CA WT and MUT cells cultured as 3D spheroids for 10 days. Clustering was performed as in Figure 1D using the median lipid intensities of 3 biological replicates. (F) Overall, precision and recall classification accuracies for PIK3CA mutation status in primary and PDX breast tumors (n = 30 in total), using all detectable lipid features (n = 1147) following 3-fold cross validation repeated 100 times with random forest as a classifier. n.s., not significant; ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001. P values in (C, bottom panel) were calculated with one-way ANOVA, followed by unpaired, two-tailed Student’s t test with Bonferroni correction.