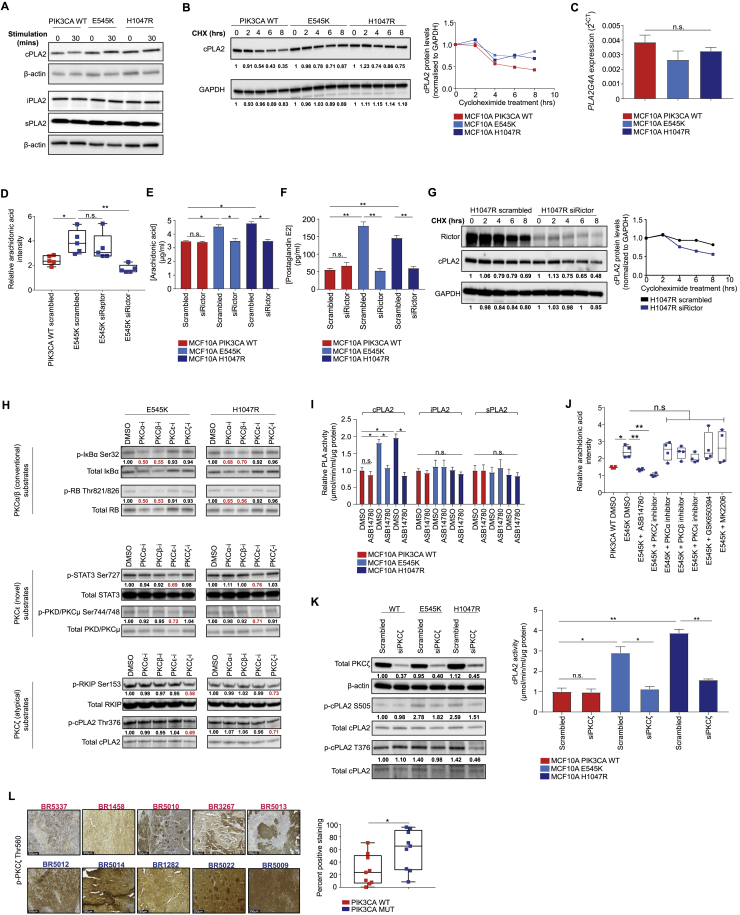
Figure S5.
Related to Figure 4
(A) Immunoblot of phospholipases in MCF10A PIK3CA WT and MUT cells following serum and growth-factor deprivation for 16 hours and stimulation with serum and growth factors for 30 min. (B) Immunoblot analysis of cPLA2 protein decay following treatment with 50 μM cycloheximide (CHX) for the indicated times. Image and quantification is from one experiment. (C) Real-time quantitative PCR of PLA2G4A expression in the MCF10A PIK3CA isogenic panel. (D) AA levels measured by REIMS in MCF10A E545K PIK3CA MUT cells following RAPTOR and RICTOR siRNA-mediated knockdown 48 hours post transfection under exogenous FAF conditions. ELISA analysis of (E) AA and (F) PGE2 in the MCF10A PIK3CA isogenic panel 48 hours post RICTOR siRNA-mediated knockdown. (G) Immunoblot analysis of cPLA2 protein decay (top) and quantification (bottom) following RICTOR siRNA-mediated knockdown and treatment with 50 μM cycloheximide for the indicated times. Image and quantification is from one experiment. (H) Immunoblot analysis of substrates of conventional PKCα/β (p-IκBα Ser32 and p-RB Thr821/826), novel PKCε (p-STAT3 Ser727 and p-PKD Ser744/748) and atypical PKCζ (p-RKIP Ser153 and p-cPLA2 T376) isoforms following treatment of MCF10A E545K and H1047R MUT cells with 1 μM of each PKCα, β, ε, and ζ peptide inhibitors for 72 hours. (I) Enzymatic activity of cPLA2, iPLA2, and sPLA2 in the MCF10A PIK3CA isogenic panel following treatment with 100 nM ASB14780 for 72 hours. (J) AA levels measured by REIMS in MCF10A E545K MUT cells treated with 100 nM ASB14780, 1 μM each of PKCα, β, ε, and ζ peptide inhibitors, 250 μM GSK650394, or 150 nM MK2206 for 72 hours under exogenous FAF conditions. (K) Immunoblot (right) of total PKCζ and phospho-S505 and T376 cPLA2 of MCF10A PIK3CA WT and MUT cells following PKCζ siRNA-mediated knockdown, and cPLA2 activity (left) following 48 hours post-transfection. (L) Representative phospho-PKCζ Thr560 immunoreactivity images (left) of 9 PIK3CA MUT (blue) and 9 WT (red) breast PDX tumors. Scale bar = 250 μm. Quantification of percent positive regions (right) was performed using the IHC profiler plug-in for ImageJ. Data are presented as the mean ± SEM of n = 3-5 biological replicates and are representative of at least two independent experiments. n.s., not significant, ∗p ≤ 0.05; ∗∗p ≤ 0.01. P values in (C), (D), (E), (F), (I), (J) and (K, right) with one-way ANOVA followed by unpaired, two-tailed Student’s t test with Bonferroni correction, and in (L) with unpaired, two-tailed Student’s t test.