FIGURE 1.
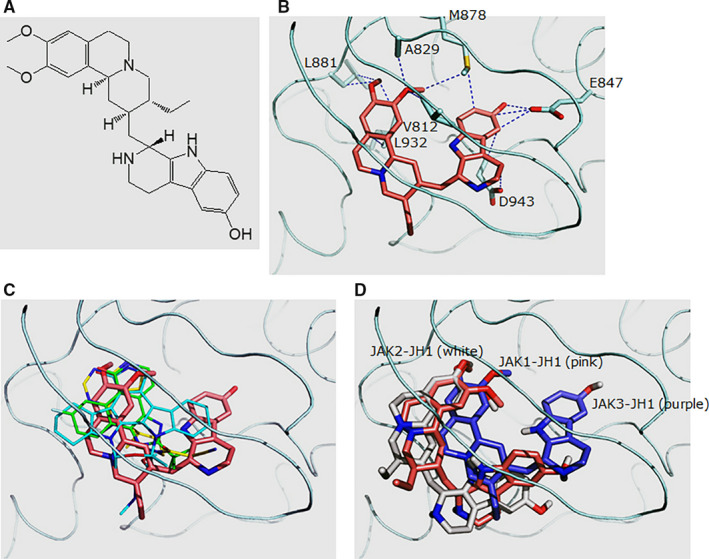
Schematic modelling of structure‐based JAK3 computational database screening. A, The chemical structure of tubulosine (C29H37N3O3). B, Predicted binding model between tubulosine and the JAK3 kinase domain (JAK3‐JH1). Tubulosine is coloured in pink. The residues that contact tubulosine with side chain atoms within 3.5 Å are labelled. C, Overlay of different ligands in complex with JAK3‐JH1. The following structures are shown: tubulosine (pink), AFN941 (cyan), CP‐690,550 (yellow) and CMP‐6 (green). These structures were generated from PDB files of 1YVJ, 24 3LXK 26 and 3LXL, 26 respectively. D, Predicted binding model between tubulosine and the kinase domains of JAK family members JAK1 (JAK1‐JH1), JAK2 (JAK2‐JH1) and JAK3 (JAK3‐JH1). JAK1‐JH1, JAK2‐JH1 and JAK3‐JH1 are coloured in pink, white and purple, respectively. All the structural figures were generated using Pymol (http://pymol.sourceforge.net/). JAK3, Janus kinase 3; JH1, Janus homology 1
