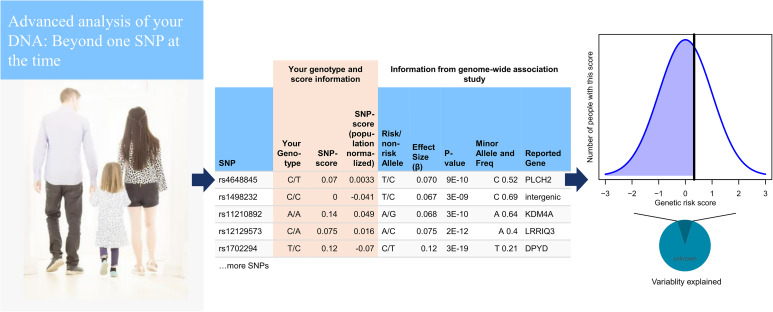
FIGURE 1.
Basic pipeline setup from the user point of view. On upload of a genome, data are checked according quality control (QC) parameters that have been developed to handle most types of microarray-based consumer genetics data. The genome is then imputed using 1000 Genomes as reference (left). The imputed data are then further subjected to automated analysis scripts from 15 different modules, most of which are based on polygenic risk score calculations. The calculations include 1,859 traits from genome-wide association studies (GWASs) and 634 traits from the UK Biobank, as well as customized modules for height, and drug response. Most polygenic risk scores use GWAS significant single-nucleotide polymorphisms (SNPs) out of necessity, although 20 major diseases are based on LDpred all-SNP scores (center). A user can then browse their scores in relation to the population, shown together with a chart displaying how much variability is explained (right).