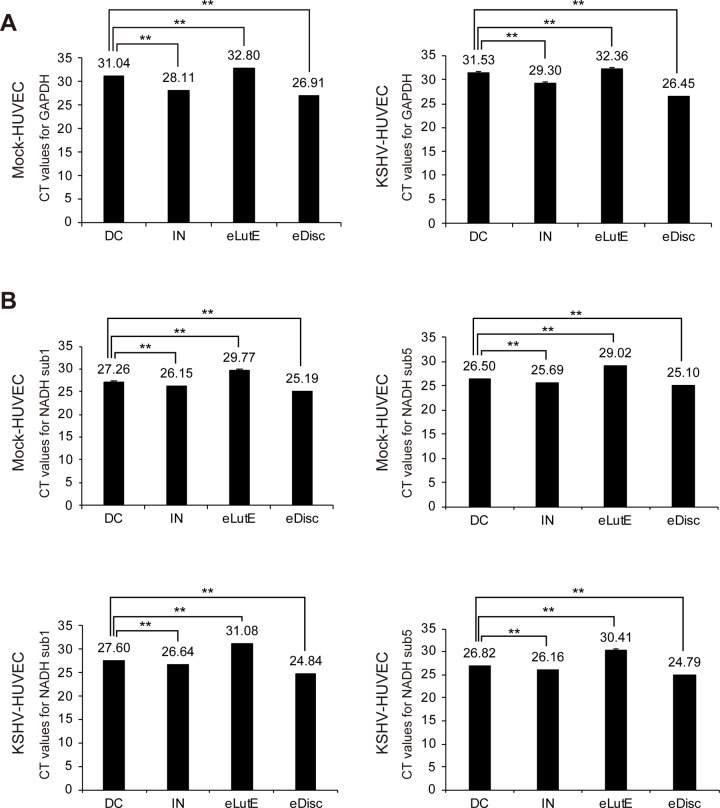
Fig 4. Analysis of DNA from EVs separated using each method.
EVs were separated from the supernatants of non-infected or KSHV-infected HUVECs using the four different EV separation methods. Equal volumes of DNA extracted from the EVs separated by each method were analyzed with qRT-PCR. qRT-PCR was used to analyze genomic and mitochondrial DNA for GAPDH (A) and NADH subunits 1 and 5 (B), respectively. The number above the bar graph indicates the average CT value from the qRT-PCR reaction. CT values from differential centrifugation were used as a control. DC: EVs separated using differential centrifugation. IN: EVs separated using the Invitrogen Total Exosome Isolation reagent. eLutE: EVs separated using the ExoLutE exosome isolation kit. eDisc: EVs separated using Exodisc from LabSpinner. Mock-HUVEC: mock-infected HUVECs. KSHV-HUVEC: KSHV-infected HUVECs. Data are shown as the mean ± SD, n = 3, **p < 0.01.