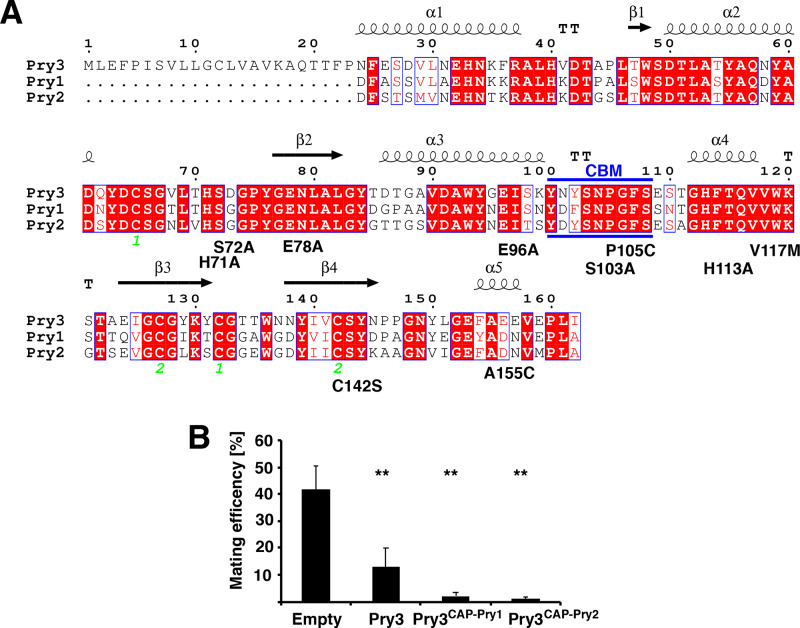
Fig. 2.
Mating inhibition is a conserved function of the yeast CAP domain. (A) The CAP domain of Pry3 is highly homologous to that of Pry1 and Pry2. Sequence alignment of the CAP domain from yeast Pry3 to that of Pry1 and Pry2 was generated with ESPrit 3.0 (Robert and Gouet, 2014). Secondary structure elements are indicated above the sequences, conserved residues are boxed red. Green numbers indicate disulfide bridges. Blue lines highlight the position of the Caveolin-Binding Motif (CBM). Mutations introduced into Pry3 CAP domain in this study are indicated below the alignment. (B) Variants of Pry3 containing the CAP domain of either Pry1 or Pry2 are functional in inhibiting mating. Mating efficiency of cells containing an empty plasmid, overexpressing a wild-type version of Pry3, or the indicated variants of Pry3 containing the CAP domain of either Pry1, Pry3CAP-Pry1 or Pry2, Pry3CAP-Pry2. Values represent means±s.d. of four independent determinations. Asterisks denote statistical significance (Welch t-test; **P-value <0.01;***P-value <0.001; n.s., not significant).