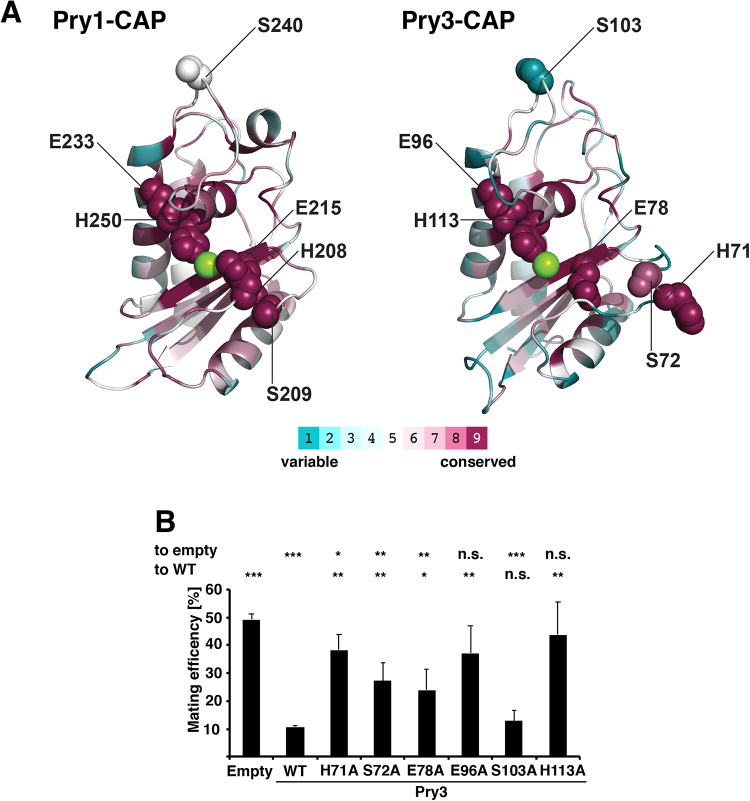
Fig. 4.
Mating inhibition by Pry3 requires putative active site residues. (A) Analysis of conserved surface residues of the CAP domain of Pry1 and Pry3. A ConSurf analysis was performed on the CAP domain of Pry1 and Pry3. Purple indicates highly conserved amino acids, whereas variable residues are in cyan as depicted in the color code bar. The central cation is indicated in green. The putative active site residues histidine 71, serine 72, glutamic acid 78, glutamic acid 96 and histidine 113, which line the central CAP cavity are highly conserved, while serine 103, which is located far away from the proposed active site shows only low conservation. (B) Conserved putative active site residues of Pry3 are required for mating inhibition. Cells containing an empty plasmid, a plasmid overexpressing wild-type Pry3, or the indicated mutant forms of Pry3 were mated to wild-type cells and mating efficiency was quantified. Values represent means±s.d. of four independent experiments. Asterisks denote statistical significance with respect to cells containing the empty plasmid or to cells overexpressing wild-type Pry3 (WT) (Welch t-test; *P-value <0.05; **P-value <0.01; ***P-value<0.001; n.s., not significant).