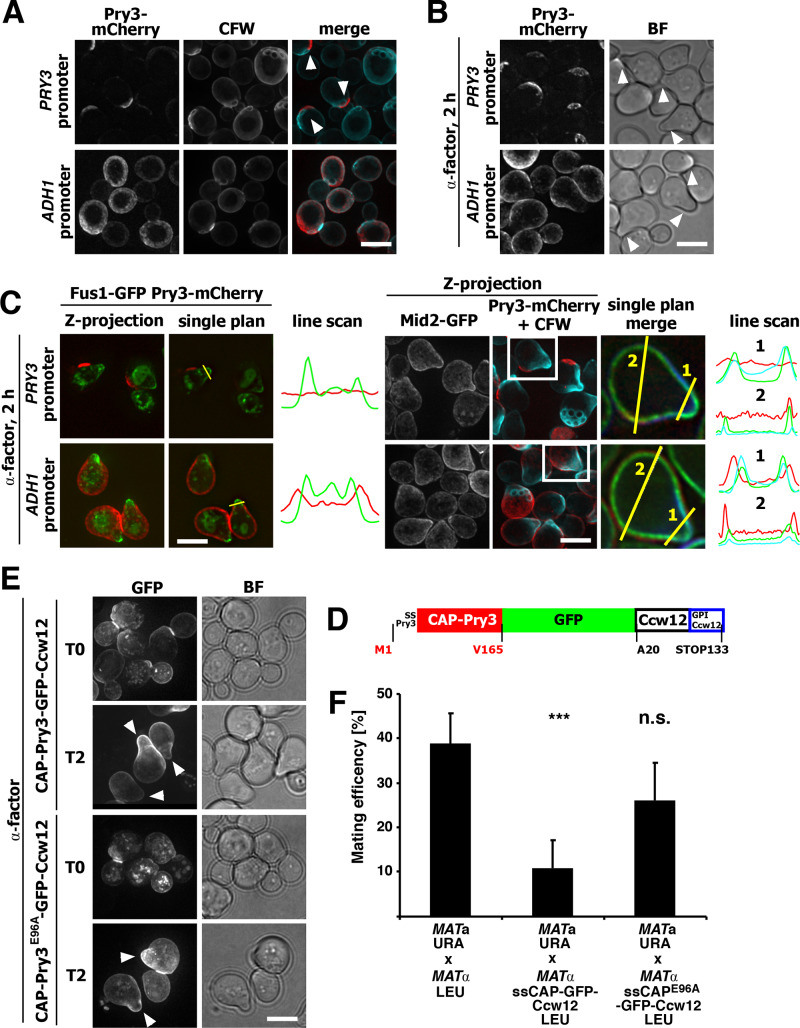
Fig. 7.
Endogenous Pry3 is absent from mating projections. (A) Pry3 localizes adjacent to bud scars. Bud scars were stained with calcofluor-white (CFW) and colocalized with mCherry-tagged Pry3 expressed from its endogenous promoter or from the ADH1 promoter. Arrowheads mark the crescent-like localization of Pry3-mCherry. (B) Pry3 localizes next to mating projections. Cells expressing mCherry-tagged Pry3 either from its endogenous promoter or from the ADH1-promoter were treated with alpha-factor for 2 h. Arrowheads point to mating projections. (C) Overexpression of Pry3 results in its mislocalization onto mating projections marked by Fus1-GFP, Mid-GFP, or CFW. Cells coexpressing Fus1-GFP or Mid2-GFP with mCherry-tagged Pry3, either from its endogenous promoter or from an ADH1 promoter, were treated with mating factor for 2 h prior to imaging. Line scans indicate the presence of Pry3-mCherry at the neck of mating projections in cells overexpressing Pry3. (D) Schematic representation of the fusion construct between the CAP domain of Pry3, GFP and the GPI-anchored cell wall protein Ccw12. The CAP domain of Pry3 is represented by the red bar, GFP by the green bar and Ccw12 by the white bar. Sites of fusions are indicated by amino acid positions. (E) The CAP-Ccw12 fusion protein localizes to mating projections. Cells expressing the wild-type or E96A mutant version of the fusion protein were treated with alpha-factor and their localization was analyzed by fluorescence microscopy. Arrowheads mark mating projections. Scale bars: 5 µm; BF, bright field illumination. (F) The CAP-Ccw12 fusion protein reduces mating efficiency. Wild-type cells were mated to cells overexpressing the fusion with a wild-type CAP domain (CAP-Ccw12) or the E96A mutant version of the CAP domain (CAPE96A-Ccw12) and mating efficiency was quantified. Values represent means±s.d. of two independent experiments. Asterisks denote statistical significance (Welch t-test; ***P-value <0.001; n.s., not significant).