Abstract
Chronic myeloid leukemia (CML) is caused by the Philadelphia (Ph+) chromosome carrying the BCR-ABL oncogene, a constitutively active tyrosine kinase. The discovery of imatinib represents a major success story in the treatment against CML. However, mutations in the BCR-ABL kinase domain are a major cause of resistance to imatinib, demonstrating that BCR-ABL remains a critical drug target. Here, we investigate a novel small molecule inhibitor, OGP46, for its inhibitory activity against K562, a panel of murine BaF3 cell lines stably expressing either wild-type BCR-ABL or its mutant forms, including T315I. OGP46 exhibits potent activity against imatinib-resistant BCR-ABL mutations, including T315I. OGP46 induced cell differentiation accompanied by G0/G1 cell-cycle arrest and suppressed the colony formation capacity of cells. Treatment with OGP46 significantly decreased the mRNA and protein expression of BCR-ABL in K562 and BaF3-p210-T315I cells. Mechanistically, the anti-cancer activity of OGP46 induced by cell differentiation is likely through the BCR-ABL/JAK-STAT pathway in native BCR-ABL and mutant BCR-ABL, including T315I, of CML cells. Our findings highlight that OGP46 is active against not only native BCR-ABL but also 11 clinically relevant BCR-ABL mutations, including T315I mutation, which are resistant to imatinib. Thus, OGP46 may be a novel strategy for overcoming imatinib-resistance BCR-ABL mutations, including T315I.
Keywords: chronic myeloid leukemia, imatinib resistance, BCR-ABL kinase, targeted therapies, cell differentiation, BCR-ABL mutations, JAK-STAT pathway, T315I mutation
Graphical Abstract

Chen and colleagues demonstrate that a small molecule, OGP46, is active against not only native BCR-ABL but also 11 clinically relevant BCR-ABL mutations, including T315I, that are resistant to imatinib by cell differentiation through the BCR-ABL/JAK-STAT pathway, which provides a novel strategy for overcoming imatinib-resistance BCR-ABL mutations in chronic myeloid leukemia.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative neoplasm characterized by the Philadelphia (Ph+) chromosome, which is caused by genetic translocation t(9; 22) (q34; q11).1,2 The reciprocal (9; 22) translocation fusion between the ABL tyrosine kinase gene on chromosome 9 and the BCR tyrosine kinase gene on chromosome 22 results in the formation of BCR-ABL fusion oncoprotein.3 The BCR-ABL fusion protein is a constitutively active protein tyrosine kinase responsible for malignant transformation by activating its downstream signaling pathways, including the JAK/STAT, phosphatidylinositol 3-kinase (PI3K)/AKT, and mitogen-activated protein kinase (MAPK)/ERK pathways, leading to aberrant cell survival.4,5 Generally, BCR-ABL is the single driving force in the pathogenesis of CML.
Imatinib (STI571) was the first BCR-ABL tyrosine kinase inhibitor (TKI) approved by the United States Food and Drug Administration for the treatment of CML.6 The introduction of imatinib has dramatically improved the outcome of patients with CML. Although imatinib can effectively treat CML, acquired resistance can be developed.7,8 Known attributors responsible for resistance to imatinib in CML are the point mutations of the BCR-ABL kinase domain,9,10 the amplification of the BCR-ABL gene,11 and the insensitivity of leukemia stem cells to imatinib.12,13 Mutation in the kinase domain of BCR-ABL is believed to be the most predominant, which gives rise to active mutant enzymes that are insensitive to imatinib.14,15 More than 70 individual mutations have been described in patients with CML.15 The most common mutations in relapsed patients are T315I, E255K, Y253F, and M351T, accounting for approximately 60% or more of all mutations that were found to be associated with imatinib resistance.16 The T315I mutant represents 15%–20% of all clinically observed mutations and is the most resilient to imatinib.17 In addition, patients with T315I mutation have a poor prognosis, with a short survival from the initiation of imatinib treatment. Over the years, second generation (nilotinib and dasatinib) and third generation (ponatinib and bosutinib) TKIs have been developed to overcome the acquired resistance. Moreover, the BCR-ABL-T315I mutation causes resistance to all first and second generation TKIs. At present, the only clinically available inhibitor that has shown efficacy against T315I mutant BCR-ABL-driven leukemia is ponatinib.18,19 However, serious adverse side effects, such as blood clots and narrowing of blood vessels, limit its clinical utility.20 Moreover, a broad spectrum of kinase inhibition increases the off-target effects of TKIs.21 Hence, identification of novel compounds and development of new strategies for the effective treatment of CML with BCR-ABL mutations, as well as T315I mutation, are important and challenging tasks.
High proliferation with an undifferentiated state is a common feature of CML.22 The CML cell line K562 behaves as a pluripotent hematopoietic precursor expressing in undifferentiated markers. K562 cells can undergo further differentiation in megakaryocytic or erythroid lineages depending on the stimulus.23 Similarly, BCR-ABL-transformed Ba/F3 cells can be induced to differentiation, and the colony formation ability of cells was reduced.24 Promoting the differentiation of leukemia cells has become a novel therapeutic strategy for the treatment of acute promyelocytic leukemia. However, cancer differentiation therapy has not been translated to the treatment of CML.
OGP46, a small molecule of kaurene diterpenoid, and its analogs have shown anti-proliferation in solid tumor cell lines, such as Eca-109, MCF-7, A549, or SMMC-7721.25, 26, 27 In this study, we investigated whether OGP46 possesses activity against leukemia cell line K562, a wide array of BaF3 cells with BCR-ABL mutations, including T315I.
Results
OGP46 Significantly Inhibits Proliferation of Leukemic Cell Lines Independent of BCR-ABL Mutational Status
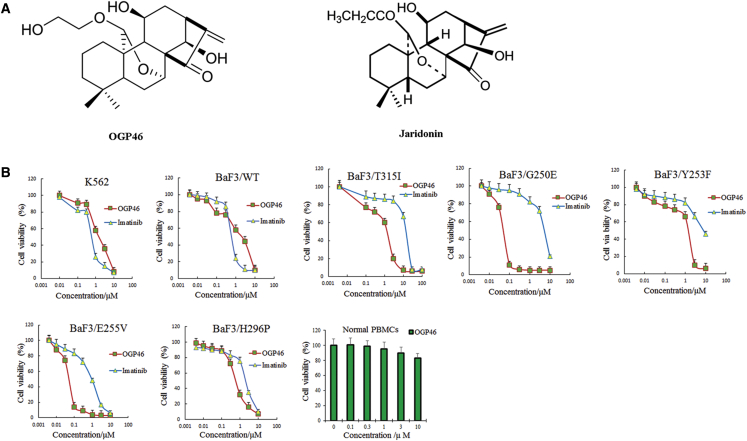
In order to test the effect of OGP46 on cell viability, we performed MTT assay. OGP46 significantly inhibited the proliferation of BaF3-p210-T315I cells as compared with imatinib (Figure 1B). The inhibition of OGP46 was about 10-fold higher than imatinib (IC50 OGP46 = 1.54 μM versus IC50 imatinib = 15.84 μM) (Table 1). This result indicated that OGP46 is effective against the imatinib-resistant T315I mutation.
Figure 1.
OGP46 Inhibits the Cell Proliferation of K562 Cells, BaF3 Cells Expressing Wild-Type BCR-ABL and Mutated BCR-ABL, and Human Normal PBMCs
(A) Chemical structure of OGP46 and Jaridonin. (B) The effect of OGP46 or imatinib on the proliferation of cells. Cells were incubated with various concentrations of OGP46 or imatinib for 72 h, then subjected to cell proliferation assay. The points with error bars represent the mean ± SD. The figures are representative of three independent experiments done in triplicate.
Table 1.
IC50 Values for OGP46 and Imatinib on K562 and BaF3 Cell Lines
| Cell Lines | OGP46, IC50 (μM) ± SD | Imatinib, IC50 (μM) ± SD |
|---|---|---|
| K562 | 1.72 ± 0.4 | 0.68 ± 0.1 |
| BaF3-p210-WT | 1.31 ± 0.3 | 0.71 ± 0.09 |
| BaF3-p210-T315I | 1.54 ± 0.8 | 15.84 ± 2.1 |
| BaF3-p210-Y253F | 1.57 ± 0.3 | 9.01 ± 0.5 |
| BaF3-p210-E255K | 0.062 ± 0.03 | 0.75 ± 0.09 |
| BaF3-p210-M351T | 1.71 ± 0.5 | 2.63 ± 0.9 |
| BaF3-p210-G250E | 0.060 ± 0.02 | 6.70 ± 0.6 |
| BaF3-p210-Q252H | 0.057 ± 0.03 | 0.38 ± 0.12 |
| BaF3-p210-E255V | 0.059 ± 0.03 | 0.85 ± 0.15 |
| BaF3-p210-F311L | 0.064 ± 0.04 | 0.21 ± 0.06 |
| BaF3-p210-H396R | 0.77 ± 0.11 | 1.01 ± 0.21 |
| BaF3-p210-H296P | 0.69 ± 0.08 | 2.19 ± 0.2 |
| BaF3-p210-M244V | 0.94 ± 0.1 | 1.71 ± 0.3 |
| BaF3-p210-F317L | 0.70 ± 0.12 | 0.090 ± 0.04 |
| BaF3-p210-F359V | 2.73 ± 0.9 | 0.44 ± 0.13 |
IC50 values obtained from cell proliferation assays. IC50 (μM) ± SD: the compound concentration that inhibited cell survival by 50% (means ± SD). Values are representative of three independent experiments, each performed in triplicate.
OGP46 also showed a similar effect on K562, BaF3-p210-wild type (WT), and another 10 mutant cell lines, including the most common mutations BaF3-p210-Y253F, BaF3-p210-M351T, and BaF3-p210-E255K. These findings reveal that OGP46 has a wide spectrum of activity on BCR-ABL mutations as compared with imatinib. However, OGP46 was less potent to BaF3-p210-F359V and BaF3-p210-F317L compared with imatinib. Interestingly, OGP46 did not have any significant effect on the viability of normal peripheral blood mononuclear cells (PBMCs).
OGP46 Induces G0/G1 Arrest in BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
In order to better explore the anti-proliferation mechanism of OGP46, we determined its effect on cell-cycle progression in BaF3-p210-WT, BaF3-p210-T315I, and K562 cells. OGP46 at 2 μM had similar effects on all of these cells. A percentage of G0/G1 constantly increased in these cell lines after treatment by OGP46 for up to 72 h (Figures 2A and 2B). These findings suggest OGP46 may inhibit cell proliferation by inducing a G0/G1 cell-cycle arrest.
Figure 2.
The Effect of OGP46 on the Cell- cycle of BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
(A) Cells were incubated with 2 μM OGP46 for 48 or 72 h, then subjected for flow cytometry analysis with PI staining. (B) The statistical analysis of cells in G0/G1 phase is presented as a bar graph (∗∗p < 0.01 versus control group). The figures are representative of three independent experiments done in triplicate.
OGP46 Induces Less Apoptosis in BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
In order to assess whether the effect of OGP46 was associated with induction of apoptosis, we incubated BaF3-p210-WT, BaF3-p210-T315I, and K562 cells with 1, 2, or 4 μM OGP46 for 72 h. The results indicated that BaF3-p210-WT did not undergo apoptosis with 1, 2, or 4 μM OGP46 (Figures 3A and 3B). Similarly, BaF3-p210-T315I and K562 cells showed no or minimal signs of apoptosis with 1 or 2 μM OGP46. However, BaF3-p210-T315I and K562 cells showed signs of apoptosis at 4 μM OGP46. In addition, it can be seen that the imatinib-resistant BaF3-p210-T315I cells did not undergo apoptosis with the imatinib treatment. These results suggest that the G0/G1 cell-cycle arrest in all of these cells induced by OGP46 at 2 μM was not due to apoptosis.
Figure 3.
The Effect of OGP46 on the Apoptosis of BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
(A) Cells were incubated with OGP46 (1–4 μM) or imatinib (2 or 4 μM) for 72 h. Cells were then subjected to flow cytometry analysis with Annexin V/PI staining. (B) The statistical analysis of apoptosis is presented as a bar graph (∗p < 0.05, ∗∗p < 0.01). The figures are representative of three independent experiments done in triplicate.
OGP46 Induces Cell Differentiation in BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
Because OGP46 induced the G0/G1 cell-cycle arrest in BaF3-p210-WT, BaF3-p210-T315I, and K562 cells without causing apoptosis, morphology analysis and flow cytometry analysis were performed to evaluate the effect of OGP46 on the differentiation in these cell lines. As shown in Figure 4A, after being treated with 2 μM OGP46 for 72 h, three cell types presented morphological changes consistent with differentiation, including polyploidization and phenotype changes (major increases in cell size accompanied by decreases in the nuclear/cytoplasmic ratio) by Wright-Giemsa staining. In contrast, the morphology of cells treated with imatinib was minimally altered. It indicates that OGP46 induces the cell differentiation by morphological changes, but imatinib does not. Moreover, it can be seen from Figures 4B and 4C that 2 μM OGP46 significantly upregulated the expression of differentiation gene F4/80 (a macrophage differentiation marker), CD25 (T cell differentiation markers), and CD61 (megakaryocytic differentiation marker) in both BaF3-p210-WT and BaF3-p210-T315I cell lines. Similarly, granulocyte differentiation was validated by the increase of CD13 expression in K562 cells incubated with 2 μM OGP46. In addition, differentiation markers CD24 and CD37 were upregulated in K562 cells. Based on these results, it can be seen that the cell-cycle exit induced by OGP46 may be caused by cell differentiation.
Figure 4.
The Effect of OGP46 on Cell Differentiation in BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
(A) Morphological features. (B and C) The expression of cell membrane markers (antigens) (B) and the expression of antigens in mean fluorescence intensity (MFI), and the statistical analysis of MFI represented as a bar graph (∗p < 0.05, ∗∗p < 0.01) (C). Cells were incubated with 2 μM OGP46 for 72 h. The figures are representative of three independent experiments. The black lines represent control, and the green lines represent OGP46.
OGP46 Significantly Reduces Colony Formation Ability in BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
We next determined the long-term efficacy of OGP46 on the formation of colonies in BaF3-p210-WT, BaF3-p210-T315I, and K562 cells. As shown in Figures 5A and 5B, the amount of colony formation was significantly reduced with increasing concentrations of OGP46. Moreover, the cells could not form colonies in either cell type when incubated with 2 μM OGP46 compared with the control group because differentiated cells have lost their abilities for colony formation. These results indicated that OGP46 could effectively suppress colony formation ability in these cells.
Figure 5.
The Effect of OGP46 on Colony Formation Ability in BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
(A) Cells were incubated with 2 μM OGP46 for 15 days and then observed by light microscopy. (B) The statistical analysis of colony formation is presented as a bar graph (∗p < 0.05, ∗∗p < 0.01). The figures are representative of three independent experiments.
OGP46 Induces Cell Differentiation by BCR-ABL Depletion
To understand the molecular consequences of cell differentiation induced by OGP46, we examined the mechanism of action of OGP46 on mRNA expression by mRNA sequencing (mRNA-seq) in BaF3-p210-WT, BaF3-p210-T315I, and K562 cells. A total of 591 genes were upregulated and 248 genes were downregulated in the K562 cell line. Similarly, the expression levels of 551 and 109 genes were increased, whereas 144 and 84 genes were decreased in BaF3-p210-WT and BaF3-p210-T315I, respectively. Therefore, there are different effects on various mRNA species, suggesting OGP46 activity does not affect all species universally. As shown in Figure 6B, cell surface antigens (F4/80 [Adgre1], CD25 [Il2rα], and CD61 [Itgb3]) were significantly upregulated in both BaF3-p210-WT and BaF3-p210-T315I cell lines. Analogously, OGP46 significantly upregulated the cell surface antigen (CD13, CD24, CD37) in the K562 cell line. Furthermore, there was an enhanced activation of cell-cycle inhibitor CDKN2A in both BaF3-p210-WT and BaF3-p210-T315I cell lines. Similarly, the expression of cell-cycle regulator CCNE2 gene was significantly reduced in the K562 cell line. In addition, cell differentiation-related CSF2RB2 and CSF2RB genes were significantly upregulated in BaF3-p210-WT, BaF3-p210-T315I, and K562 cells, respectively. As shown in Figure 6C, by Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis in BaF3-p210-WT, BaF3-p210-T315I, and K562 cells, several signaling pathways were involved in the OGP46 treatment, such as JAK-STAT or pathways in cancer, hematopoietic linage, and cell adhesion molecular pathways, which all are involved in regulation of cell differentiation. Additionally, the antigen processing and presentation signal pathway was involved in regulation of immunology. The signal pathways consistently demonstrated several such related genes (CD25 [JAK-STAT]; BCR-ABL, CD25 (pathways in cancer); CD13, CD24, CD25, CD61, CSF2RB2, and CSF2RB [hematopoietic cell lineage]; and CD40 [cell adhesion molecules]).
Figure 6.
The Action of OGP46 Is Related to the JAK-STAT Pathway to Promote Cell Differentiation in BaF3-p210-WT, BaF3-p210-T315I, and K562 Cells
(A) Volcano plots. (B) Heatmap of differentially expressed genes (62 genes are the common differentially expressed genes of BaF3-p210-WT and BaF3-p210-T315I; 100 genes according to their p value in the K562 cell line) and SP1, BCR, and MYC genes. (C) KEGG pathway analyses of all differentially expressed genes. Cells were incubated with 2 μM OGP46 for 48 h; then mRNA sequencing was performed. The figures are representative of two independent experiments. ∗p < 0.05 versus control group.
To validate the differentially expressed genes identified by mRNA-seq, we performed real-time PCR and western blotting in BaF3-p210-T315I and K562 cell lines. As shown in Figures 7A and 7C, the mRNA expression and protein of CCNE2 and CDKN2A were significantly altered, which was in accordance with the mRNA-seq result.
Figure 7.
OGP46 Induces Cell Differentiation by BCR-ABL Inhibition in K562 and BaF3-p210-T315I Cells
(A) The effects of OGP46 on the mRNA expression were determined by real-time PCR (n = 3) in both cell lines. Cells were incubated with 2 μM OGP46 for 0, 24, 48, or 72 h. (B) BCR-ABL mRNA expression in BaF3-p210-T315I and K562 cells treated with 2 μM OGP46, or actinomycin D (4 μM) for 0, 6, 12, 24 or 48 h. The mRNA expression is reported as percentage of total transcript present at the time 0. (C) BCR-ABL mRNA expression in both cell lines treated with actinomycin D (4 μM), or the combination of 2 μM OGP46 with actinomycin D (4 μM) for 0, 6, 12, or 24 h. (D) The effects of OGP46 on the protein levels were determined by western blotting (n = 3) in both cell lines. Cells were incubated with OGP46 (0.5–2 μM) or imatinib (1 or 2 μM) for 72 h. (E) Protein expression was quantified by the software AI600 images. ∗∗p < 0.01 versus control group.
Because BCR-ABL is a critical fusion gene that causes CML clonal evolution, we next examined whether OGP46 inhibited BCR-ABL at the transcriptional and protein levels in both cell lines. It was shown that BCR-ABL was inhibited by OGP46 at both levels, whereas imatinib had no effect on native BCR-ABL and BCR-ABL T315I mutation cells (Figures 7A–7E). These results indicate that the sensitivity of native BCR-ABL and BCR-ABL-T315I mutation cells to OGP46 might be due to the inhibition of BCR-ABL expression. In contrast, sensitivity of native BCR-ABL cells to imatinib is due to the p-BCR-ABL, which inhibits the activity of BCR-ABL. However, imatinib cannot affect either the BCR-ABL nor the p-BCR-ABL in BCR-ABL-T315I mutation cells.
To investigate when BCR-ABL mRNA decreases and how rapidly after treatment with OGP46, stability of BCR-ABL mRNA due to OGP46 treatment was compared with the effect of a broad transcriptional inhibitor actinomycin D (ActD) in BaF3-p210-T315I and K562 cell lines. The data showed that OGP46 did not significantly decrease the BCR-ABL mRNA in 12 h. As shown in Figure 7B, BCR-ABL mRNA was decreased by 52% in 6 h after treatment with ActD, while a similar amount of BCR-ABL mRNA was decreased in about 20 h after treatment with OGP46. Moreover, as shown in Figure 7C, OGP46 did not lead to further decreases in BCR-ABL mRNA levels beyond that seen by ActD alone. Therefore, the action of OGP46 is different from ActD, suggesting that OGP46 uniquely affects BCR-ABL mRNA transcription rather than globally affects multiple species or mRNA stability.
Discussion
CML will become the most prevalent form of leukemia in the next 20–30 years for increased patient survival. Since its introduction in 2001, imatinib represents a major success in the treatment of CML in targeted cancer therapy.28 However, about 50% of patients experience intolerance or resistance to imatinib. The most common mechanism of treatment failure is the acquisition of mutations in the kinase domain of BCR-ABL.29 Among them, T315I point mutation are the most common, and afflicted patients often result in poor prognosis. This led to the development of second generation and third generation TKIs. Ponatinib, a third generation TKI, has been demonstrated to be effective against the highly T315I mutation; however, serious adverse side effects were observed in patients.30,31 Moreover, the off-target effects of TKIs is increased by a broad spectrum of kinase inhibition. Therefore, resistance to TKIs due to BCR-ABL mutation, including T315I, remains a major challenge for patients with CML.32 In the present study, we aimed to develop a novel compound to overcome drug resistance induced by BCR-ABL mutations, including T315I, in CML.
OGP46 has a similar chemical structure with a natural ent-kaurenoid diterpenoid Jaridonin, which induces esophageal cancer cells apoptosis. OGP46 and Jaridonin have the same maternal structure, the difference is the substituent. Here, we discovered that OGP46 potently inhibited proliferation of leukemic cell lines independent of BCR-ABL mutational status and induced cell differentiation in CML cells harboring native BCR-ABL or BCR-ABL-T315I mutation accompanied by cell-cycle arrest at G0/G1. OGP46 dramatically downregulated the mRNA and protein of BCR-ABL regardless of the mutational status of BCR-ABL. All together, we found that OGP46 is effective against native BCR-ABL-positive CML cells and a panel of BaF3 cells bearing BCR-ABL mutants, including T315I, by cell differentiation through depletion of BCR-ABL.
CML is one of the typical models of myeloproliferative disorders characterized by the presence of BCR-ABL.33 It is well documented that the BCR-ABL fusion protein blocks the differentiation of CML cells and protects the cells from apoptosis.34 Hence inducing CML cell differentiation represents a promising therapeutic strategy in the treatment of CML.35,36 It is reported that As4S4 can induce effective erythroid differentiation through degradation of BCR-ABL.34 Furthermore, silencing BCR-ABL in the lineage leads to enhanced erythroid differentiation and a decrease in colony-forming capacity in CML patient samples.37 Here, we found that OGP46 had an anti-proliferative effect on native BCR-ABL CML cells, a panel of CML cells bearing BCR-ABL mutants, as well as T315I mutation by inducing cell differentiation. Meanwhile, the mRNA and protein of BCR-ABL were depleted by OGP46. These data indicate that the cell differentiation induced by OGP46 is originated by inhibition of the expression of BCR-ABL mRNA. Compared with ActD, the results showed that OGP46 does not inhibit mRNA globally nor affect mRNA stability (Figures 6B, 7B, and 7C). Rather, it decreases the expression of BCR-ABL mRNA by inhibiting the transcription of BCR-ABL evidence by that OGP46 does not act additively with ActD to decrease BCR-ABL mRNA level in K562 and BaF3-p210-T315I cells (Figure 7C). Furthermore, JAK-STAT, a downstream signaling pathway of BCR-ABL, has been proved to be related to cell differentiation.38,39 Hence the inhibition of BCR-ABL by OGP46 resulting in cell differentiation might be involved in the JAK-STAT signaling pathway in BaF3-p210-WT and BaF3-p210-T315I (Figure 6C). Consistently, we found that BCR-ABL depletion led to cell differentiation that might be involved in pathways in cancer signaling pathway in the K562 cell line (Figure 6C). Moreover, the pathways in cancer signaling pathway are involved in the BCR-ABL and JAK-STAT signaling pathway (KEGG map; https://www.genome.jp/kegg-bin/show_pathway?hsa05200). Therefore, OGP46 is effective against native or mutant BCR-ABL, including T315I, in CML cell lines, likely through BCR-ABL/JAK-STAT signaling pathways.
Differentiation of cells is usually accompanied by cell-cycle exit.40 The onset of differentiation that occurs in G1 phase is concomitant with cell-cycle exit and can be mediated and maintained by the following: (1) upregulation of CDK inhibitor proteins; and (2) downregulation of CCNE2, CCND1, and CCNE1.41 For example, the CDK inhibitor, such as CDKN2A, was upregulated, whereas CCNE2 was downregulated in the differentiation response. Our results show that OGP46 significantly inhibited cell proliferation by inducing cell differentiation accompanied by cell-cycle arrest at G0/G1 (Figure 2). The cell-cycle arrest at G0/G1 was associated with activation of CDKN2A or suppression of CCNE2 identified by mRNA-seq (Figure 6B) and confirmed by PCR and western blot (Figures 7A, 7D, and 7E).
Taken together, our results suggest that the small molecule inhibitor OGP46 is effective against imatinib-resistance BCR-ABL mutations, including T315I, in CML. Moreover, our findings demonstrate that OGP46 targets native and mutant BCR-ABL, including T315I, in CML cells through BCR-ABL/JAK-STAT signaling pathways, which subsequently results in cell differentiation (Figure 8). Interestingly, OGP46 does not significantly affect the viability of normal PBMCs. Its ability to overcome imatinib resistance induced by either T315I or other major resistance mutations means OGP46 could be a promising lead compound that merits further development to overcome imatinib-resistance BCR-ABL mutations, including T315I, in CML.
Figure 8.
Model of Anti-cancer Effects of OGP46 in CML
In general, imatinib can effectively treat CML caused by BCR-ABL oncoprotein by inhibition of the activity of BCR-ABL (A). However, mutations in the BCR-ABL kinase domain, including T315I, are a major cause of resistance to imatinib (B). We newly found that OGP46 is effective against not only native BCR-ABL but also imatinib-resistant BCR-ABL mutations, including T315I (C), by cell differentiation through the BCR-ABL/JAK-STAT pathway. Cell differentiation is accompanied by cell-cycle arrest at G0/G1, potentiating anti-cancer effects.
Materials and Methods
Chemicals and Equipment
OGP46 was prepared from Dr. Hong-Min Liu’s lab (Zhengzhou University, China). The molecular weight of OGP46 [C22H32O6 with the nomenclature 20(S)-11β, 14β-dihydroxy-20-(2-hydroxyethoxy)-7α, 20-epoxy-ent-kaur-16-en-15-one] is 392.49, and its purity was more than 98.5%. Stock solutions of OGP46 (10 mM; dissolved in dimethyl sulfoxide) were stored at −20°C. The same concentration of DMSO in RPMI 1640 medium was used as vehicle in various in vitro assays. Figure 1A shows the chemical structure of OGP46 and Jaridonin.26 Imatinib mesylate was purchased from TSZ Chem (Lexington, MA, USA). RPMI-1640, fetal bovine serum (FBS), 5,000 U/mL penicillin, and 5,000 μg/mL streptomycin were purchased from GIBCO (Carlsbad, CA, USA). Propidium iodide (PI)/RNase staining buffer and the Fluorescein Isothiocyanate (FITC) Annexin V Apoptosis Detection Kit were purchased from BD Biosciences (San Jose, CA, USA). FITC anti-human CD13 (Cat #11-0138-42, RRID: AB_11043278), phycoerythrin (PE) anti-mouse CD25 (Cat #56-0251-60, RRID: AB_891424), and PE anti-mouse CD61 (Cat #13-0611-81, RRID: AB_466487) antibodies were purchased from eBioscience (San Diego, CA, USA). PE anti-human CD24 (Cat #561646) and PE anti-human CD37 (Cat #561546) antibodies were purchased from BD Biosciences (San Jose, CA, USA). FITC anti-mouse F4/80 (Cat #60027FI.1) antibody and MethoCult H4100 (Cat #04100) were purchased from STEMCELL Technologies (Vancouver, BC, Canada). Antibodies against BCR-ABL (Cat #3902), pBCR-ABL (Cat #3901), and GAPDH (Cat #5174) were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies against CDKN2A (ab211542) and CCNE2 (ab32103) were purchased from Abcam (Cambridge, MA, USA). The PrimerScript RT reagent kit and the SYBR Premix Ex Taq reagent kit were purchased from TAKARA Bio (Otsu, Japan). Flow cytometry analyses were conducted using a FACSCalibur System (BD Biosciences, San Diego, CA, USA). PCR amplification was performed using an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA).
Cell Lines and Cell Culture
Human cell line K562 and murine BaF3 cells expressing WT BCR-ABL (BaF3-p210-WT) and BCR-ABL single mutants at each of the 13 key positions (BaF3-p210-T315I, BaF3-p210-G250E, BaF3-p210-E255V, BaF3-p210-F359V, BaF3-p210-H296P, BaF3-p210-M315T, BaF3-p210-Y253F, BaF3-p210-Q252H, BaF3-p210-H396R, BaF3-p210-F311L, BaF3-p210-M244V, BaF3-p210-F317L, and BaF3-p210-E255K) were provided by Dr. Zhe-Sheng Chen’s lab (St. John’s University, USA). Cell lines expressing WT BCR-ABL or BCR-ABL with various kinase domain point mutations were derived by transfection of a retroviral vector expressing p210BCR-ABL into murine hematopoietic cells as described previously.42 Cord blood samples from three healthy individuals (obtained from The Affiliated Hospital of Weifang Medical University, Weifang, China) were collected after obtaining written informed consent from the donor. The PBMCs were isolated using a Histopaque 1077 by gradient centrifugation. All cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin.
Cell Proliferation Assay
The anti-proliferative effects of OGP46 were determined by a modified MTT colorimetric assay. 5 × 103 cells per well were seeded into a 96-well plate. After 24 h of incubation, the cells were treated with either OGP46 or imatinib at the indicated concentrations. After 72 h, 20 μL MTT (4 mg/mL) reagent was added to each well, and the cells were further incubated at 37°C for 4 h. Following incubation, the plates were centrifuged, and the formazan crystals were dissolved in 100 μL DMSO. The absorbance was measured at 570 nm by an Opsys microplate reader (Dynex Technologies, Chantilly, VA, USA).
Cell-Cycle Analysis
Cells were incubated with 2 μM OGP46 for different time intervals (0, 48, or 72 h). The cells were collected at the end of each time interval. The cells were fixed by 100% cold ethanol and subsequently stained with 50 μg/mL PI and 100 μg/mL RNase A for 1 h at room temperature in the dark. Flow cytometry analysis was used to determine the percentage of cells in a particular phase of the cell cycle with a BD Accuri C6 flow cytometer (San Jose, CA, USA).
Annexin V/PI Analysis
To determine apoptotic cells, we incubated cells with 1, 2, or 4 μM OGP46 for 72 h. The cells were collected, washed with PBS, resuspended in the binding buffer, and incubated with FITC-labeled Annexin V and PI (BD Biosciences) for 30 min at room temperature in the dark. The apoptotic cell population was determined by flow cytometry analysis.
Cell Morphology Analysis
Cells were cultured with a specified concentration of OGP46 for 72 h. The cells were collected, and slides were made and fixed by methanol for 30 min, stained with Wright-Giemsa, and observed for morphological features of differentiation by light microscopy.
Cell Surface Antigens Assessment
Cells were cultured with a specified concentration of OGP46. After 72 h of being cultured, the cells were collected, washed with PBS, and incubated with different monoclonal antibodies. The cell surface antigens were evaluated by flow cytometry.
Colony Formation Assay
Cells (5 × 104) were cultured with OGP46 in 500 μL of 2.6% methylcellulose medium containing 10% FBS in 24-well plates. After incubation for 15 days, the number of separate colonies was counted by an inverted microscope.
mRNA-Seq
mRNA-seq was performed for K562, BaF3-p210-WT, and BaF3-p210-T315I cell lines (Annoroad Gene Technology, Beijing, China). In brief, RNA was extracted, and the quality was assessed. 2 μg of total RNA was purified and converted to an Illumina sequencing library, and the libraries were validated. Sequencing was performed with an Illumina HiSeq 4000 platform, and 150-bp paired-end reads were generated. Sequencing reads were mapped to the genome using TopHat version 2.1.1. The expressions of mRNAs were estimated using transcripts per million (TPMs). Differential expression test was analyzed using DESeq R packages according to the package’s manual. The p values were adjusted, and a corrected p value of 0.05 was set as a threshold for significant differential expression of genes. Gene Ontology (GO) function and KEGG enrichment analyses of differently expressed genes (DEGs) were performed with R language with the aid of packages clusterProfiler, enrichplot, and ggplot2. Terms with a p value of <0.05 were considered significantly enriched.
Quantitative Real-Time PCR
Total RNA was extracted using the TRIzol Reagent (Invitrogen Life Technologies). The cDNA was transcribed from 1 μg total RNA using a PrimerScript RT reagent kit with a gDNA Eraser. Quantitative real-time PCR was performed in triplicate using SYBR Green I and hotstart Taq DNA polymerase in an Applied Biosystems 7500 Fast Real-Time PCR System. The relative quantification of gene expression was calculated using the 2−ΔΔCt method. The following primers were used: forward BCR-ABL (mouse) 5′-TTCAGAAGCTTCTCCCTGACAT-3′, reverse BCR-ABL (mouse) 5′-CTTCGTCTGAGATACTGGATTCCT-3′; forward CDKN2A (mouse) 5′-TTGGCCCAAGAGCGGGGACA-3′, reverse CDKN2A (mouse) 5′-GCGGGCTGAGGCCGGATTTA-3′; forward BCR-ABL (human) 5′-TCCGCTGACCATCAATAAGGA-3′, reverse BCR-ABL (human) 5′-CACTCAGACCCTGAGGCTCAA-3′; and forward CCNE2 (human) 5′-CTTACGTCACTGATGGTGCTTGC-3′, reverse CCNE2 (human) 5′-CTTGGAGAAAGAGATTTAGCCAGG-3′.
Western Blotting Analysis
Equal amounts of cell lysate were electrophoresed onto a sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membrane. The membranes were blocked and then incubated with antibodies. Images were obtained using an enhanced chemiluminescence reagent (ECL) detection system (Amersham Imager 600; GE Healthcare Biosciences, Pittsburgh, PA, USA).
Statistical Analysis
All experiments were repeated at least three times unless otherwise stated. Statistical significance between the control group and each treatment group was determined by ANOVA. Statistical significance level was p <0.05 or p <0.01.
Author Contributions
L.W.: methodology, performed experiments, dada analysis, and writing – original draft preparation. Y.Y.: performed PCR and western blotting analysis. P.G.: performed a major part of MTT assay. M.Z.: prepared study materials. M.Q.: prepared the chemical reagent solutions. Y.Z.: provision of instrumentation. Y.K.: prepared the compound OGP46 studied in this paper. Y.L.: prepared the compound OGP46 studied in this paper. H.-M.L.: provided the compound OGP46 studied in this paper. X.X.: analyzed study data. A.W. and Y.S.: collected the cord blood samples of healthy donors for PBMC isolation. Z.-S.C.: supervision, conceptualization, and writing – reviewing and editing. Z.H.: supervision, conceptualization, and writing – reviewing and editing.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81370628 and 81570157 to Z.H.), National Natural Science Foundation of China (grant 81700167), and Natural Science Foundation of Shandong Province (grant ZR2016HM47 to L.W.).
Contributor Information
Zhe-Sheng Chen, Email: chenz@stjohns.edu.
Zhenbo Hu, Email: huzhenbo@wfmc.edu.cn.
References
- 1.Rowley J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Schuster C., Forster K., Dierks H., Elsässer A., Behre G., Simon N., Danhauser-Riedl S., Hallek M., Warmuth M. The effects of Bcr-Abl on C/EBP transcription-factor regulation and neutrophilic differentiation are reversed by the Abl kinase inhibitor imatinib mesylate. Blood. 2003;101:655–663. doi: 10.1182/blood-2002-01-0043. [DOI] [PubMed] [Google Scholar]
- 3.Bartram C.R., de Klein A., Hagemeijer A., van Agthoven T., Geurts van Kessel A., Bootsma D., Grosveld G., Ferguson-Smith M.A., Davies T., Stone M. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–280. doi: 10.1038/306277a0. [DOI] [PubMed] [Google Scholar]
- 4.Lugo T.G., Pendergast A.M., Muller A.J., Witte O.N. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 5.Danial N.N., Rothman P. JAK-STAT signaling activated by Abl oncogenes. Oncogene. 2000;19:2523–2531. doi: 10.1038/sj.onc.1203484. [DOI] [PubMed] [Google Scholar]
- 6.Talpaz M., Silver R.T., Druker B.J., Goldman J.M., Gambacorti-Passerini C., Guilhot F., Schiffer C.A., Fischer T., Deininger M.W., Lennard A.L. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a phase 2 study. Blood. 2002;99:1928–1937. doi: 10.1182/blood.v99.6.1928. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien S.G., Guilhot F., Larson R.A., Gathmann I., Baccarani M., Cervantes F., Cornelissen J.J., Fischer T., Hochhaus A., Hughes T., IRIS Investigators Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 8.Hughes T., Saglio G., Branford S., Soverini S., Kim D.W., Müller M.C., Martinelli G., Cortes J., Beppu L., Gottardi E. Impact of baseline BCR-ABL mutations on response to nilotinib in patients with chronic myeloid leukemia in chronic phase. J. Clin. Oncol. 2009;27:4204–4210. doi: 10.1200/JCO.2009.21.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah N.P. Loss of response to imatinib: mechanisms and management. Hematology (Am. Soc. Hematol. Educ. Program) 2005;1:183–187. doi: 10.1182/asheducation-2005.1.183. [DOI] [PubMed] [Google Scholar]
- 10.Shah N.P., Nicoll J.M., Nagar B., Gorre M.E., Paquette R.L., Kuriyan J., Sawyers C.L. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 11.Gambacorti-Passerini C.B., Gunby R.H., Piazza R., Galietta A., Rostagno R., Scapozza L. Molecular mechanisms of resistance to imatinib in Philadelphia-chromosome-positive leukaemias. Lancet Oncol. 2003;4:75–85. doi: 10.1016/s1470-2045(03)00979-3. [DOI] [PubMed] [Google Scholar]
- 12.Corbin A.S., Agarwal A., Loriaux M., Cortes J., Deininger M.W., Druker B.J. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J. Clin. Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham S.A., Hopcroft L.E., Carrick E., Drotar M.E., Dunn K., Williamson A.J., Korfi K., Baquero P., Park L.E., Scott M.T. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apperley J.F. Chronic myeloid leukaemia. Lancet. 2015;385:1447–1459. doi: 10.1016/S0140-6736(13)62120-0. [DOI] [PubMed] [Google Scholar]
- 15.Gontarewicz A., Balabanov S., Keller G., Colombo R., Graziano A., Pesenti E., Benten D., Bokemeyer C., Fiedler W., Moll J., Brümmendorf T.H. Simultaneous targeting of Aurora kinases and Bcr-Abl kinase by the small molecule inhibitor PHA-739358 is effective against imatinib-resistant BCR-ABL mutations including T315I. Blood. 2008;111:4355–4364. doi: 10.1182/blood-2007-09-113175. [DOI] [PubMed] [Google Scholar]
- 16.La Rosée P., Corbin A.S., Stoffregen E.P., Deininger M.W., Druker B.J. Activity of the Bcr-Abl kinase inhibitor PD180970 against clinically relevant Bcr-Abl isoforms that cause resistance to imatinib mesylate (Gleevec, STI571) Cancer Res. 2002;62:7149–7153. [PubMed] [Google Scholar]
- 17.Redaelli S., Piazza R., Rostagno R., Magistroni V., Perini P., Marega M., Gambacorti-Passerini C., Boschelli F. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J. Clin. Oncol. 2009;27:469–471. doi: 10.1200/JCO.2008.19.8853. [DOI] [PubMed] [Google Scholar]
- 18.Cortes J.E., Kim D.-W., Pinilla-Ibarz J., le Coutre P., Paquette R., Chuah C., Nicolini F.E., Apperley J.F., Khoury H.J., Talpaz M., PACE Investigators A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2013;369:1783–1796. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Hare T., Shakespeare W.C., Zhu X., Eide C.A., Rivera V.M., Wang F., Adrian L.T., Zhou T., Huang W.S., Xu Q. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zabriskie M.S., Eide C.A., Tantravahi S.K., Vellore N.A., Estrada J., Nicolini F.E., Khoury H.J., Larson R.A., Konopleva M., Cortes J.E. BCR-ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell. 2014;26:428–442. doi: 10.1016/j.ccr.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mian A.A., Rafiei A., Haberbosch I., Zeifman A., Titov I., Stroylov V., Metodieva A., Stroganov O., Novikov F., Brill B. PF-114, a potent and selective inhibitor of native and mutated BCR/ABL is active against Philadelphia chromosome-positive (Ph+) leukemias harboring the T315I mutation. Leukemia. 2015;29:1104–1114. doi: 10.1038/leu.2014.326. [DOI] [PubMed] [Google Scholar]
- 22.Faderl S., Talpaz M., Estrov Z., O’Brien S., Kurzrock R., Kantarjian H.M. The biology of chronic myeloid leukemia. N. Engl. J. Med. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- 23.Jacquel A., Herrant M., Defamie V., Belhacene N., Colosetti P., Marchetti S., Legros L., Deckert M., Mari B., Cassuto J.P. A survey of the signaling pathways involved in megakaryocytic differentiation of the human K562 leukemia cell line by molecular and c-DNA array analysis. Oncogene. 2006;25:781–794. doi: 10.1038/sj.onc.1209119. [DOI] [PubMed] [Google Scholar]
- 24.Neviani P., Santhanam R., Trotta R., Notari M., Blaser B.W., Liu S., Mao H., Chang J.S., Galietta A., Uttam A. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8:355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Ke Y., Wang W., Zhao L.F., Liang J.J., Liu Y., Zhang X., Feng K., Liu H.M. Design, synthesis and biological mechanisms research on 1,2,3-triazole derivatives of Jiyuan Oridonin A. Bioorg. Med. Chem. 2018;26:4761–4773. doi: 10.1016/j.bmc.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y.C., Ke Y., Zi X., Zhao W., Shi X.J., Liu H.M. Jaridonin, a novel ent-kaurene diterpenoid from Isodon rubescens, inducing apoptosis via production of reactive oxygen species in esophageal cancer cells. Curr. Cancer Drug Targets. 2013;13:611–624. doi: 10.2174/15680096113139990030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ke Y., Liang J.J., Hou R.J., Li M.M., Zhao L.F., Wang W., Liu Y., Xie H., Yang R.H., Hu T.X. Synthesis and biological evaluation of novel Jiyuan Oridonin A-1,2,3-triazole-azole derivatives as antiproliferative agents. Eur. J. Med. Chem. 2018;157:1249–1263. doi: 10.1016/j.ejmech.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 28.Kantarjian H.M., Talpaz M., Giles F., O’Brien S., Cortes J. New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann. Intern. Med. 2006;145:913–923. doi: 10.7326/0003-4819-145-12-200612190-00008. [DOI] [PubMed] [Google Scholar]
- 29.Gorre M.E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P.N., Sawyers C.L. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 30.Valent P., Hadzijusufovic E., Schernthaner G.H., Wolf D., Rea D., le Coutre P. Vascular safety issues in CML patients treated with BCR/ABL1 kinase inhibitors. Blood. 2015;125:901–906. doi: 10.1182/blood-2014-09-594432. [DOI] [PubMed] [Google Scholar]
- 31.Cortes J.E., Kantarjian H., Shah N.P., Bixby D., Mauro M.J., Flinn I., O’Hare T., Hu S., Narasimhan N.I., Rivera V.M. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2012;367:2075–2088. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Bubnoff N., Schneller F., Peschel C., Duyster J. BCR-ABL gene mutations in relation to clinical resistance of Philadelphia-chromosome-positive leukaemia to STI571: a prospective study. Lancet. 2002;359:487–491. doi: 10.1016/S0140-6736(02)07679-1. [DOI] [PubMed] [Google Scholar]
- 33.Sawyers C.L. Shifting paradigms: the seeds of oncogene addiction. Nat. Med. 2009;15:1158–1161. doi: 10.1038/nm1009-1158. [DOI] [PubMed] [Google Scholar]
- 34.Wang T., Wen T., Li H., Han B., Hao S., Wang C., Ma Q., Meng J., Liu J., Xu H. Arsenic sulfide nanoformulation induces erythroid differentiation in chronic myeloid leukemia cells through degradation of BCR-ABL. Int. J. Nanomedicine. 2019;14:5581–5594. doi: 10.2147/IJN.S207298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacquel A., Colosetti P., Grosso S., Belhacene N., Puissant A., Marchetti S., Breittmayer J.P., Auberger P. Apoptosis and erythroid differentiation triggered by Bcr-Abl inhibitors in CML cell lines are fully distinguishable processes that exhibit different sensitivity to caspase inhibition. Oncogene. 2007;26:2445–2458. doi: 10.1038/sj.onc.1210034. [DOI] [PubMed] [Google Scholar]
- 36.Mazharian A., Ghevaert C., Zhang L., Massberg S., Watson S.P. Dasatinib enhances megakaryocyte differentiation but inhibits platelet formation. Blood. 2011;117:5198–5206. doi: 10.1182/blood-2010-12-326850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rangatia J., Bonnet D. Transient or long-term silencing of BCR-ABL alone induces cell cycle and proliferation arrest, apoptosis and differentiation. Leukemia. 2006;20:68–76. doi: 10.1038/sj.leu.2403999. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda T., Nakamura T., Nakao K., Arai T., Katsuki M., Heike T., Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai H., Qin X., Yang C. Dehydrocostus Lactone Suppresses Proliferation of Human Chronic Myeloid Leukemia Cells Through Bcr/Abl-JAK/STAT Signaling Pathways. J. Cell. Biochem. 2017;118:3381–3390. doi: 10.1002/jcb.25994. [DOI] [PubMed] [Google Scholar]
- 40.Hass R., Gunji H., Datta R., Kharbanda S., Hartmann A., Weichselbaum R., Kufe D. Differentiation and retrodifferentiation of human myeloid leukemia cells is associated with reversible induction of cell cycle-regulatory genes. Cancer Res. 1992;52:1445–1450. [PubMed] [Google Scholar]
- 41.Gendelman R., Xing H., Mirzoeva O.K., Sarde P., Curtis C., Feiler H.S., McDonagh P., Gray J.W., Khalil I., Korn W.M. Bayesian Network Inference Modeling Identifies TRIB1 as a Novel Regulator of Cell-Cycle Progression and Survival in Cancer Cells. Cancer Res. 2017;77:1575–1585. doi: 10.1158/0008-5472.CAN-16-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta P., Kathawala R.J., Wei L., Wang F., Wang X., Druker B.J., Fu L.W., Chen Z.S. PBA2, a novel inhibitor of imatinib-resistant BCR-ABL T315I mutation in chronic myeloid leukemia. Cancer Lett. 2016;383:220–229. doi: 10.1016/j.canlet.2016.09.025. [DOI] [PubMed] [Google Scholar]