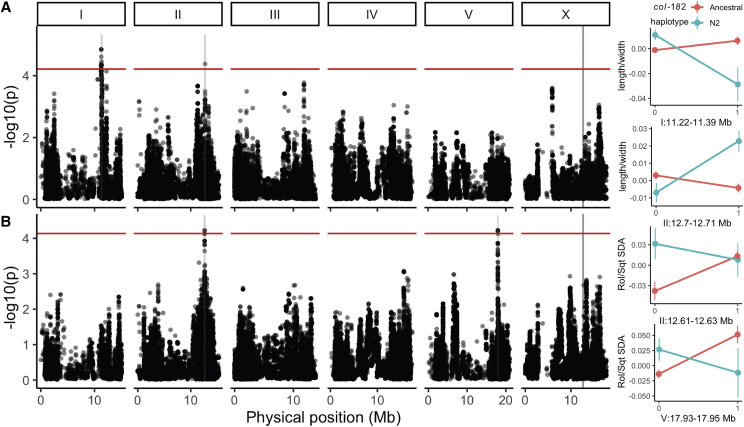
Figure 4.
col-182 modifies the effects of natural genetic variation on worm movement and shape. Genome-wide statistics are shown for two bivariate response models of col-182 indel SNP genotype, worm length/width (A) and Rol/Sqt sparse discriminant functions (B), using diallelic SNPs segregating in 363 recombinant inbred lines of the C. elegans multiparent experimental evolution (CeMEE) panel. Statistics are from a likelihood ratio test for a full additive and interaction linear model against a null model of no genetic effects. Permutation thresholds for genome-wide significance (FDR = 0.2) are shown in red, light shaded regions show 1.5 LOD drop QTL intervals (expanded to a minimum of 300 Kb for visibility), and the location of col-182 on the X chromosome is indicated by a dark gray line. Effect plots at right show genotype class means and standard errors for QTL with significant interactions (parametric bootstrap against additive models, ). Trait values shown are the first principal component for length/width, and the Sqt discriminant function, which explains most of the interaction in the Rol/Sqt bivariate model. Reference-based genotype is on the x-axis. Genotype and phenotype data in File S15, code in File S16.