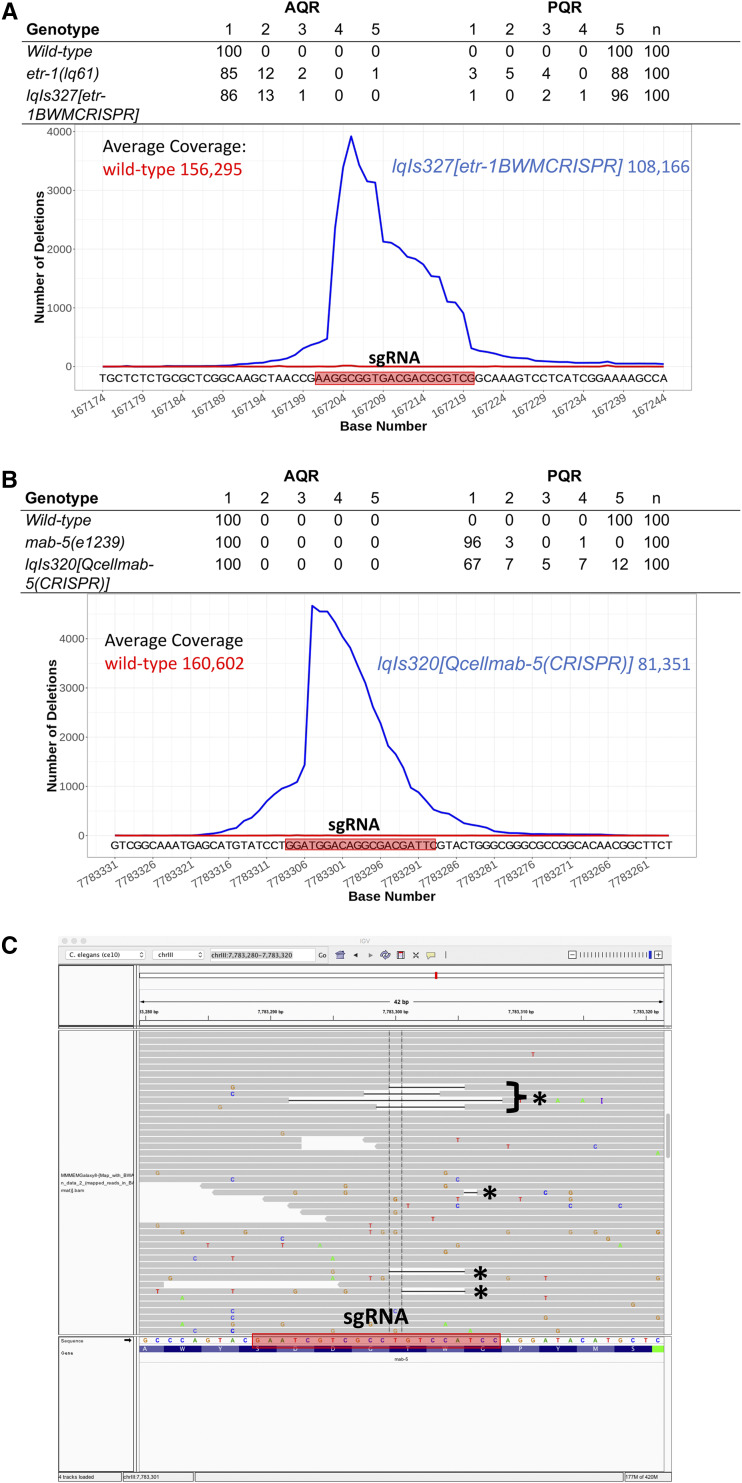
Figure 6.
CRISPR-seq of etr-1 and mab-5 cell-specific genome editing. A) The table shows AQR and PQR migration in wild-type, an etr-1(lq61) mutant, and a transgenic etr-1(CRISPR) animal with ubiquitous etr-1 sgRNA expression and Cas9 expression from the myo-3 promoter expressed in body wall muscle. The graph shows the results of an amplicon sequencing experiment, with the chromosome position in nucleotides (X-axis) and the number of deletions at each position in wild-type (red) and etr-1(CRISPR) (blue) (Y-axis). The sgRNA sequence used is indicated in the red box. The average coverage in amplicon sequencing experiment across the region is indicated. B) A CRISPR-seq experiment on cell-specific mab-5 CRISPR, as described for etr-1 in (A). The mab-5 sgRNA was expressed ubiquitously, and Cas9 was expressed from the Q-cell-specific egl-17 promoter. C) A screenshot from Integrated Genome Viewer showing alignment of mab-5 CRISPR-seq reads. Reads with deletions are indicated with asterisks. The sgRNA sequence is in the red box, and is the reverse complementary strand to the site depicted in Figure 6B.