Abstract
Anopheles sinensis is a major malaria vector in Southeast Asia. Resistance to pyrethroid insecticides in this species has impeded malaria control in the region. Previous studies found that An. sinensis populations from Yunnan Province, China were highly resistant to deltamethrin and did not carry mutations in the voltage-gated sodium channel gene that cause knockdown resistance. In this study, we tested the hypothesis that other genomic variants are associated with the resistance phenotype. Using paired-end whole genome sequencing (DNA-seq), we generated 108 Gb of DNA sequence from deltamethrin -resistant and -susceptible mosquito pools with an average coverage of 83.3× depth. Using a stringent filtering method, we identified a total of 916,926 single nucleotide variants (SNVs), including 32,240 non-synonymous mutations. A total of 958 SNVs differed significantly in allele frequency between deltamethrin -resistant and -susceptible mosquitoes. Of these, 43 SNVs were present within 37 genes that code for immunity, detoxification, cuticular, and odorant proteins. A subset of 12 SNVs were randomly selected for genotyping of individual mosquitoes by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and showed consistent allele frequencies with the pooled DNA-seq derived allele frequencies. In addition, copy number variations (CNVs) were detected in 56 genes, including 33 that contained amplification alleles and 23 that contained deletion alleles in resistant mosquitoes compared to susceptible mosquitoes. The genomic variants described here provide a useful resource for future studies on the genetic mechanism of insecticide resistance in this important malaria vector species.
Keywords: Anopheles sinensis, whole genome sequencing, insecticide resistance, genomic variant, copy number variation, polymerase chain reaction-restriction fragment length polymorphism
Malaria is one of the most important vector-borne diseases in Southeast Asia, and the Anopheles sinensis mosquito is the predominant malaria vector. Currently, insecticide-treated bed nets (ITNs) and indoor residual spraying (IRS) are the most important means of mosquito vector control in the global strategy for malaria control (WHO 2019). Pyrethroid are the only class of insecticides approved for use on ITNs due to their high toxicity to insects, rapid rate of knockdown, strong mosquito excito-repellency, and low mammalian toxicity (Diabate et al. 2002; Hemingway et al. 2004; Kaneko 2011). Extensive use of insecticides has resulted in resistance in many vector mosquito species, including malaria vectors (WHO 2012). The evolution and spread of resistance to insecticides have significantly hampered the efficacy of ITN programs (Alout et al. 2014; Coetzee and Koekemoer 2013; Srivastava et al. 2010). Tools for early detection of insecticide resistance and resistance surveillance are critical to resistance management and to the rational use of insecticides (WHO 2012).
Development of molecular diagnostic tools for insecticide resistance requires knowledge of resistance mechanisms. In vector mosquito species, at least three mechanisms of physiological resistance to pyrethroids are known (WHO 2012): 1) knockdown resistance (kdr) caused by point mutations in the pyrethroid target site, the para sodium channel gene, 2) biochemical resistance conferred by metabolic detoxification enzymes such as cytochrome P450 monooxygenases, glutathione S-transferases and esterases, and 3) penetration resistance caused by cuticular thickening. kdr mutations and their association with the resistant phenotype have been well studied in many vector species, and markers to monitor resistance have been developed (Martinez-Torres et al. 1998; Ranson et al. 2000). For example, two of the most common mutations at the same position lead to a change of a Leucine to a Phenylalanine (L1014F) or to a Serine (L1014S) in kdr gene are known to confer knockdown resistance to pyrethroids and DDT insecticides in An. gambiae s.l. (Platt et al. 2015; Lynd et al. 2018; Reimer et al. 2008; Mitchell et al. 2014; Chandre et al. 1999) in Africa. However, recent studies support the hypothesis that the kdr allele is not fully predictive of the resistant phenotype (Weetman and Donnelly 2015; Donnelly et al. 2009; Gnanguenon et al. 2015; Okorie et al. 2015; Chang et al. 2014), and resistance is likely caused by multiple genetic factors (Brooke 2008; Toé et al. 2015; Zhong et al. 2013). Gene copy number variations (CNVs) have been reported as additional mechanisms of insecticide resistance in Anopheles mosquitoes (Djogbénou et al. 2008; Lucas et al. 2019; Weetman et al. 2018a). Highly differentiated copy number variations between population samples were also reported from pooled population sequencing in Drosophila melanogaster (Schrider et al. 2013) as well as Aedes aegypti mosquitoes (Matthews et al. 2018).
Whole genome sequencing approaches have proven to be a more powerful tool for achieving a holistic understanding of insecticide resistance mechanisms than classic individual gene-based approaches (Toé et al. 2015; Zhu et al. 2014; Faucon et al. 2015; David et al. 2014). To investigate whether different genomic variants other than kdr are associated with the resistance phenotype, we compared the genome variation between deltamethrin -resistant and -susceptible mosquitoes from Yunnan, China where the An. sinensis mosquito populations lack the kdr mutations. First, we developed a comprehensive list of genetic variants in the resistant and susceptible pools of An. sinensis. Then, we examined a subset of 12 single nucleotide variants (SNVs) in an additional 40 individuals (20 resistant and 20 susceptible) using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) methods to validate the DNA-seq derived allele frequencies. Finally, we examined the CNVs between deltamethrin -resistant and -susceptible mosquitoes.
Methods And Materials
Sample collection and bioassay
In May 2012, An. sinensis mosquito larvae and pupae were collected from irrigated rice fields using standard 350ml plastic water dippers in Yinjiang County, Yunnan Province, China. Coordinates for the collection site are latitude 24°45’22.7”N and longitude 98°05’16.4”E, and the elevation is ∼850m. Mosquito larvae and pupae were collected in a variety of habitats with a maximum of 5 individuals per breeding habitat across three villages separated by ∼10 km from each other to avoid sampling siblings. This sampling scheme would yield no more than 2 female adults per habitats based on the assumption of larval-to-adult survivorship 60–80% (Afrane et al. 2007) and sex ratio of 1:1 (Phasomkusolsil et al. 2011). The collected mosquito larvae and pupae were transported to a local laboratory and reared into adults. All adult mosquitoes were identified to species using the published morphological keys of Dong (Dong 2010). Adult mosquitoes were provided with fresh 10% sucrose solution daily. Adults reared from field-collected larvae and pupae were used in insecticide bioassays to minimize the influence of mosquito age and blood feeding history on resistance measurements. After the mosquitoes were identified to species, An. sinensis female adult mosquitoes at 3–5 days post emergence were tested for susceptibility to deltamethrin using the standard WHO resistance tube bioassay (WHO 2013). Briefly, twenty-five mosquitoes were exposed to 0.05% deltamethrin-impregnated paper in an upright plastic tube for 1 h, then transferred into holding tubes and fed on 10% sugar solution on cotton wool for 24 hr. After the 24 hr recovery period, the dead (classified as susceptible) and live mosquitoes (classified as resistant) were preserved individually in 1.5ml tubes containing 1.0 ml 95% ethanol for subsequent DNA extraction and molecular identification of species. A total of 200 female mosquitoes in 8 tubes were tested for resistance against deltamethrin with an additional group of 50 individuals in 2 tubes (without insecticide exposure) as the control group. After the resistance bioassay was completed, all of the insecticide-exposed mosquitoes were preserved and used for subsequent DNA extraction, whereas the 50 living mosquitoes in control tubes were killed and discarded.
Mosquito DNA extraction and whole genome sequencing
Mosquito DNA was extracted from single mosquitoes using the QIAamp DNA Mini Kit (Qiagen Inc. Valencia, CA) according to the manufacturer’s instructions. The extracted DNA was further cleaned and concentrated using DNA Clean & Concentrator (Zymo Research, Irvine, CA). DNA samples were quantified with Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies, Carlsbad, CA) and checked for quality by 1.0% agarose gel electrophoresis. All mosquitoes were genotyped for species identification by An. sinensis allele specific PCR (AS-PCR) following a previously published protocol (Joshi et al. 2010). High-quality DNA with equal amounts at the same concentration from each of the 20 resistant mosquitoes were pooled and named YLR (resistant). Similarly, equal amounts of high-quality DNA at the same concentration from each of the 20 susceptible mosquitoes were pooled and named YLS (susceptible). The two DNA pools were used to build paired-end libraries for whole genome DNA sequencing and sequenced on the Illumina Genome Analyzer IIx (GAIIx) at the Broad Institute of MIT and Harvard by running two lanes of PE100 sequencing (100-bp paired-end reads) per pool. Bases were called using Illumina software and data outputted as fastq files.
Validation of SNV allele frequencies determined by pooled DNA sequencing
To validate the allele frequencies determined by pooled DNA sequencing in the two groups, we examined the allele frequencies by genotyping individual mosquitoes from resistant and susceptible groups. Twenty additional individuals were randomly selected from each of the phenotyped resistant and susceptible groups at the same time as those individuals used for pooled sequencing. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis was performed for the 40 additional phenotyped individuals at 12 SNVs in 12 randomly selected candidate genes. Gene-specific primers were designed using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/). In order to guarantee gene-specificity and avoid amplification of multigene families, primers were designed upon target regions which included the SNVs anchoring outside the conserved region. PCR amplifications were performed in a total volume of 20 μl with 5-20 ng genomic DNA from resistant and susceptible individuals, 10 pmol of forward and reverse primers each with SYBR Green PCR Master Mix (Life Technologies, Grand Island, NY) under the following thermocycling conditions: 95° for 3 min, then 35 cycles of 95° for 30 s, 55° for 30 s, 72° for 45 s, and finally 72° for 6 min. 5 μl of PCR products were used for restriction enzyme digestions (Table S1). After digestion, the products were run on a 2% agarose gel, with undigested PCR product as control. The mutation frequencies were calculated based on the agarose gel electrophoresis bands.
Data analysis
CLC Genomics Workbench 12.0.3 software (CLCbio, Aahus, Denmark, http://www.clcbio.com) was used for data analyses. First, Trimmomatic 0.36 (Bolger et al. 2014) was used to remove adapters and perform a sliding window of trimming to discard sequences with a Phred score of less than 30. Then, reads were filtered based on their quality using the NCBI/Sanger or Illumina pipeline function to trim low-quality reads and filter out failed reads in CLC (Leaché et al. 2013). The resulting high-quality paired-end reads from YLR pool, YLS pool, and YLR+YLS (combined reads from both pools) were separately mapped to the An. sinensis reference genome (VectorBase, www.vectorbase.org: Anopheles sinensis, AsinC2) using the default parameters. SNVs were called in the mapped sequencing reads of the YLR+YLS using the ‘Low Frequency Variant Detection’ tool of CLC against the reference genome of An. sinensis (AsinC2). The following parameters were used: required significance = 1%, ignore positions with coverage above = 800, ignore broken pairs = yes (broken paired reads defined as one of the two reads shorter than the set length cutoff at 100-bp after quality trimming), minimum coverage = 80 (40 individuals with at least 2 reads for each individual), minimum count = 28, minimum frequency = 35%, base quality filter = yes, neighborhood radius = 5, minimum central quality = 20, minimum neighborhood quality = 15, read direction filter = yes, direction frequency = 5%, forward/reverse balance >0.25. Pyro-error variants in homopolymer regions with a minimum length of 3 and a frequency below 0.8 were removed. Detection of synonymous and non-synonymous polymorphism was completed by the “Amino Acid Changes” tool within the CLC using variant file resulted from CLC and CDS track files extracted from reference genome. The WEGO software was used to display functional classification of Gene Ontology (GO) (Ye et al. 2006).
To identify SNVs in coding regions that differ in allele frequency between resistant and susceptible mosquitoes, variant calling of non-synonymous polymorphism sites was performed on the read mapping from YLR pool and YLS pool, respectively, using a track file of known variants (identified above) as input and the “Identify Known Mutations from mappings” tool within the CLC. The parameters used to filter variants were as follows: minimum coverage = 40 (20 individuals with at least 2 reads for each individual, given a minimum sequence depth > 1), detection frequency = 2.5% (1/40 alleles for detection of singletons), ignore broken pairs = yes, ignore non-specific matches = yes, and create individual tracks = yes. The resulting information from each pool was exported from the CLC to Excel format, which included the variant frequency, read count, read coverage, and other statistics of each variant locus in the read mapping of the two pooled samples. Allele frequency estimates were calculated as the fraction of reads carrying the non-reference alleles or read counts divided by read coverages. Fisher’s exact tests were used to examine the differences of variant allele frequencies between resistant and susceptible samples using the SNP_tools package (Chen et al. 2009). Frequency distribution histograms of genome-wide variants were produced using JMP Pro 14.0.0 (SAS Institute Inc., Cary, NC). Allele frequencies were considered significantly different between resistant and susceptible samples (hereafter named as differential variant) if the false discovery rate (FDR) adjusted p-value < 0.05 (Benjamini and Hochberg 1995). A sample size of 20 individuals (diploid) in each pool could detect differences larger than 30% in allele frequencies as significant, with a power of 80% at confidence interval of 95% (p < 0.05, two-tailed) (Cohen 1988). In order to identify polymorphisms most strongly associated with deltamethrin resistance, we further filtered SNVs for the candidate SNVs in which the absolute allele frequency difference was > 35% (Faucon et al. 2015). Copy number variation (CNV) detection of coding regions in deltamethrin -resistant mosquitoes was conducted by CLC using the deltamethrin -susceptible mosquitoes as control against the reference genome of An. sinensis. The statistic threshold for significance was set at an FDR adjusted p-value < 0.05 for multiple testing (Benjamini and Hochberg 1995) with a low coverage cutoff at 40.
Data availability
The data sets supporting the results of this article are available in the Sequence Read Archive under the accession number SRR830401 and SRR830336 for the Illumina whole genome sequencing of genomic DNA in resistant and susceptible mosquitoes in An. sinensis. Supplemental material available at figshare: https://doi.org/10.25387/g3.10315898
Results
Mosquito susceptibility bioassay and whole-genome sequencing
Among the 200 female mosquitoes exposed to insecticide for testing of susceptibility to deltamethrin, 132 individuals were identified as resistant and 64 as susceptible to deltamethrin (mortality rate 32.0%). All mosquitoes were identified as An. sinensis by the AS-PCR method. A total of 194,546,524 and 192,880,116 paired-end reads were obtained for deltamethrin-resistant (YLR pool) and -susceptible (YLS pool) samples, respectively. Of the total, 183,041,352 (94.09%) and 181,355,527 (94.02%) reads were mapped to the An. sinensis reference genome (AsinC2.1) for YLR pool and YLS pool respectively, resulting in a mean coverage depth of 83.7× and 83.0×. When the reads from the two pooled samples were combined, a total of 364,676,689 reads were mapped to the reference genome, resulting in a mean pooled coverage depth of 166.8× and individual genome coverage depth of 2.1× (Table 1).
Table 1. Summary of whole-genome sequencing in pyrethroid resistant (YLR pool) and susceptible (YLS pool) Anopheles sinensis.
| Deltamethrin resistant (YLR) | Deltamethrin susceptible (YLS) | Combined (YLR+YLS) | |
|---|---|---|---|
| Total number of reads | 194,546,524 | 192,880,116 | 387,426,640 |
| Mapped reads | 183,041,352 | 181,355,527 | 364,676,689 |
| % mapped reads | 94.09% | 94.02% | 94.13% |
| Average length of reads in pairs | 163.44 | 158.84 | 161.18 |
| Broken paired reads | 25,068,830 | 25,987,373 | 51,043,207 |
| Number of bases mapped | 18,487,176,552 | 18,316,908,227 | 36,832,345,589 |
| Sequencing depth | 83.7× | 83.1× | 166.8× |
Single nucleotide variants in Anopheles sinensis genome
A total of 916,926 SNVs were identified from all the sequence reads (YLR pool and YLS pool combined) against the An. sinensis reference genome. Of these, the majority of SNVs (884,686, 96.5%) were located within non-coding regions. Only 32,240 variants (3.5%) were located within coding regions and resulted in amino acid changes. Compared to the reference genome, variants at 5,418 sites within coding regions were fixed in allele frequency at 100% in both the resistant and susceptible pool, and so these sites were filtered out. The remaining 26,822 SNVs were used as variant track (target sites) for identifying SNVs from read mapping of YLR pool and YLS pool, respectively.
After we filtered out the low coverage (< 40 reads) SNVs, a total of 16,340 target SNV sites were analyzed for allele frequency differences between the two pools (Table S2). The distributions of variant frequencies in the YLR pool and YLS pool are presented in Figure 1. The frequencies were not distributed normally in both pools (KSL goodness-of-fit test: D = 0.08, p < 0.01 for YLS, and D = 0.07, p < 0.01 for YLR). An allele frequency comparison of the 16,340 SNVs between YLR pool and YLS pool led to the identification of 958 differential SNVs at an FDR adjusted p-value ≤ 0.05 for multiple testing (Figure 2A, Table S2). The distribution of frequency differences between the two pools at 16,340 SNV sites showed the goodness of fit to a normal distribution with a mean of 0.11 and a standard deviation of 14.05 (Figure 2A), whereas the frequency differences of the 958 differential SNVs clearly fit a mixture of two normal distributions (π1 = 0.50, π2 = 0.49; Figure 2B), indicating two groups of samples. The number of SNVs in the two groups (those with higher frequency in resistant and those with lower frequency in resistant pool) were approximately equal. These differential variants were distributed across 790 genes, including immunity, detoxification, cuticular, and odorant proteins at an average mutation rate of 1.6 ± 0.06 per kb (Figure 3, Table S3). More than half of the genes (54.81%, 433/790) were functionally unannotated and ∼40% of these genes were annotated with unknown functions. Among the remaining (4.68%, 37/790) genes of known function, a significant enrichment was detected in the classes of immunity (16) and detoxification proteins (14), followed by odorant proteins (5), and cuticular proteins (2). A total of 43 SNVs were present within these 37 genes. Among the 790 genes with differential variants, 351 were assigned for 1009 GO accession numbers and were classified into 37 function categories under three major domains (biological process, cellular component, and molecular function) (Figure 4, Table S4). Two molecular functions (catalytic activity and binding) and biological processes (metabolic process and cellular process) were highly enriched.
Figure 1.
Frequency distribution of single nucleotide variants (SNVs) detected in pyrethroid resistant and susceptible Anopheles sinensis. The x-axis represents SNV allele frequency. 0%, all alleles in the pool are reference; 100%, all alleles in the pool are non-reference. The y-axis represents read coverage for SNVs. A: susceptible mosquitoes (YLS pool); B: resistant mosquitoes (YLR pool). In the upper box, 25th, 50th and 75th quartiles are displayed; the mean and the 95% confidence interval are represented by a diamond.
Figure 2.
Distribution of allele frequency differences between YLS and YLR pools in Anopheles sinensis. A: Distribution of frequency difference between the two pools at 16,340 SNV sites; and B: Distribution of frequency difference between the two pools at the 958 differential SNVs (FDR adjusted p-value < 0.05). A negative value indicates high allele frequency in susceptible mosquito pool (YLS), whereas a positive value indicates high allele frequency in resistant mosquito pool (YLR).
Figure 3.
Pie charts show the number and the percentage of genes harboring the differential SNVs detected in pyrethroid resistant and susceptible Anopheles sinensis. The function of 37 annotated genes is indicated.
Figure 4.
Histogram of Gene Ontology (GO) classification of the 790 genes harboring differential genomic variants in Anopheles sinensis. Three main ontologies of GO (biological process, cellular component and molecular function) are shown in the x-axis. The left y-axis indicates the percentage of total genes and the right y-axis is the number of genes in each category.
The voltage-gated sodium channel is the target of pyrethroids insecticides. Resistance to pyrethroids is often associated with point mutations in the associated gene, which causes target site insensitivity. After carefully examining the voltage-gated sodium channel gene (GenBank acc. KFB44005, 2138 amino acids), we identified only one non-synonymous mutation (resulting in Ala1072Thr substitute) with a similar allele frequency in deltamethrin -susceptible and -resistant mosquitoes, suggesting that this amino acid change is not related to resistance. The single amino acid change Leu119Phe of glutathione S-transferase gene (GSTe2) has been reported to confer high levels of metabolic resistance to DDT in the malaria vector An. funestus (Riveron et al. 2014). We examined the variants in this gene in An. sinensis (GenBank acc. KFB39338, 221 amino acids) and found two mutations at nucleotide position 355 and 457, resulting in Leu119Val and Ala153Pro amino acid substitutions. However, for both alleles, there was no statistically significant difference in allele frequency between susceptible and resistant mosquitoes, suggesting that this gene is not related to deltamethrin resistance in this population. High organophosphate resistance resulting from insensitive acetylcholinesterase (AChE) by a single mutation (G119S of the ace-1 gene) has been reported in Culex pipiens and in An. gambiae (Weill et al. 2004). We identified three mutations at positions 239, 269, and 781 (GenBank acc. KFB35326, 385aa), which resulted in Gly80Ala, Val90Ala, and Gly261Ser (corresponding G119S of ace-1 gene in other species reported) in amino acid substitutions, but with similar allele frequencies in susceptible and resistant mosquitoes. Further study is needed to assess the association between these mutations and organophosphate resistance.
Candidate genes with the largest difference in allele frequency and high quality
Among the 790 differential genes, 88 (11.1%) genes with high quality (Phred quality score > 35) showed the largest difference (> 40%) in allele frequency between resistant and susceptible pools (FDR adjusted p-value < 0.01). These candidates included genes in the classes of immunity (ASIC011903: Met11Ile and ASIC021092: Asn365Asp), detoxification (ASIC012065: Ala1059Thr and ASIC016833: Lys495Thr), and cuticular protein (ASIC010236: Arg25Cys). The candidate genes that contained the 12 most highly differentiated SNVs between YLS and YLR pools are listed in Table 2. Among these genes, only four had gene function descriptions, including two genes (WW domain-binding protein 1 and short-chain dehydrogenase) with a high SNV allele frequency in susceptible mosquitoes and two genes (indolepyruvate ferredoxin oxidoreductase subunit alpha and putative L-carnitine dehydratase/alpha-methylacyl-CoA racemase) with a high SNV allele frequency in resistant mosquitoes.
Table 2. List of highly differential SNPs identified between YLS and YLR pools in Anopheles sinensis.
| Transcript ID | GenBank accession | Size (bp) | Gene description | Nucleic acid change | Amino acid change | Allele frequency (%, YLS) | Allele frequency (%, YLR) | Adjusted p-value |
|---|---|---|---|---|---|---|---|---|
| YLS > YLR | ||||||||
| ASIC003949 | KFB36789 | 3897 | WW domain-containing protein 1 | 2644A > G | Thr882Ala | 66.7 | 14.0 | 6.17E-04 |
| ASIC004261 | KFB37064 | 1236 | hypothetical protein ZHAS_00004261 | 159G > A | Met53Ile | 69.6 | 16.0 | 2.46E-04 |
| ASIC008461 | KFB40921 | 7764 | AGAP002735-PA-like protein | 2275C > A | His759Asn | 70.8 | 15.0 | 2.75E-04 |
| ASIC021070 | KFB52802 | 432 | hypothetical protein ZHAS_00021070 | 283G > T | Gly95Cys | 71.4 | 20.8 | 5.57E-04 |
| ASIC015363 | KFB47421 | 1479 | hypothetical protein ZHAS_00015363 | 1459T > C | Cys487Arg | 77.6 | 27.5 | 5.50E-04 |
| ASIC001969 | KFB35388 | 183 | short-chain dehydrogenase | 43C > T | Arg15Trp | 86.4 | 23.3 | 1.33E-05 |
| ASIC002752 | KFB35824 | 3462 | AGAP002548-PA-like protein | 1081T > C | Phe361Leu | 86.4 | 25.6 | 3.34E-05 |
| YLR > YLS | ||||||||
| ASIC009989 | KFB42304 | 2247 | hypothetical protein ZHAS_00009989 | 976C > T | Pro326Ser | 23.4 | 77.5 | 5.64E-04 |
| ASIC011867 | KFB44037 | 333 | indolepyruvate ferredoxin oxidoreductase subunit alpha | 252T > G | Asp84Glu | 25.0 | 79.5 | 4.63E-04 |
| ASIC021208 | KFB52925 | 4215 | AGAP005789-PA-like protein | 1354T > G | Leu452Val | 40.0 | 100 | 6.33E-06 |
| ASIC010615 | KFB42862 | 444 | AGAP008279-PA-like protein | 185C > T | Thr62Met | 44.4 | 95.2 | 2.74E-04 |
| ASIC003791 | KFB36595 | 492 | putative L-carnitine dehydratase/alpha-methylacyl-CoA racemase | 280T > A | Ser94Thr | 47.6 | 97.7 | 2.61E-04 |
Relationship between pooled DNA-seq and individually genotyping determined allele frequencies
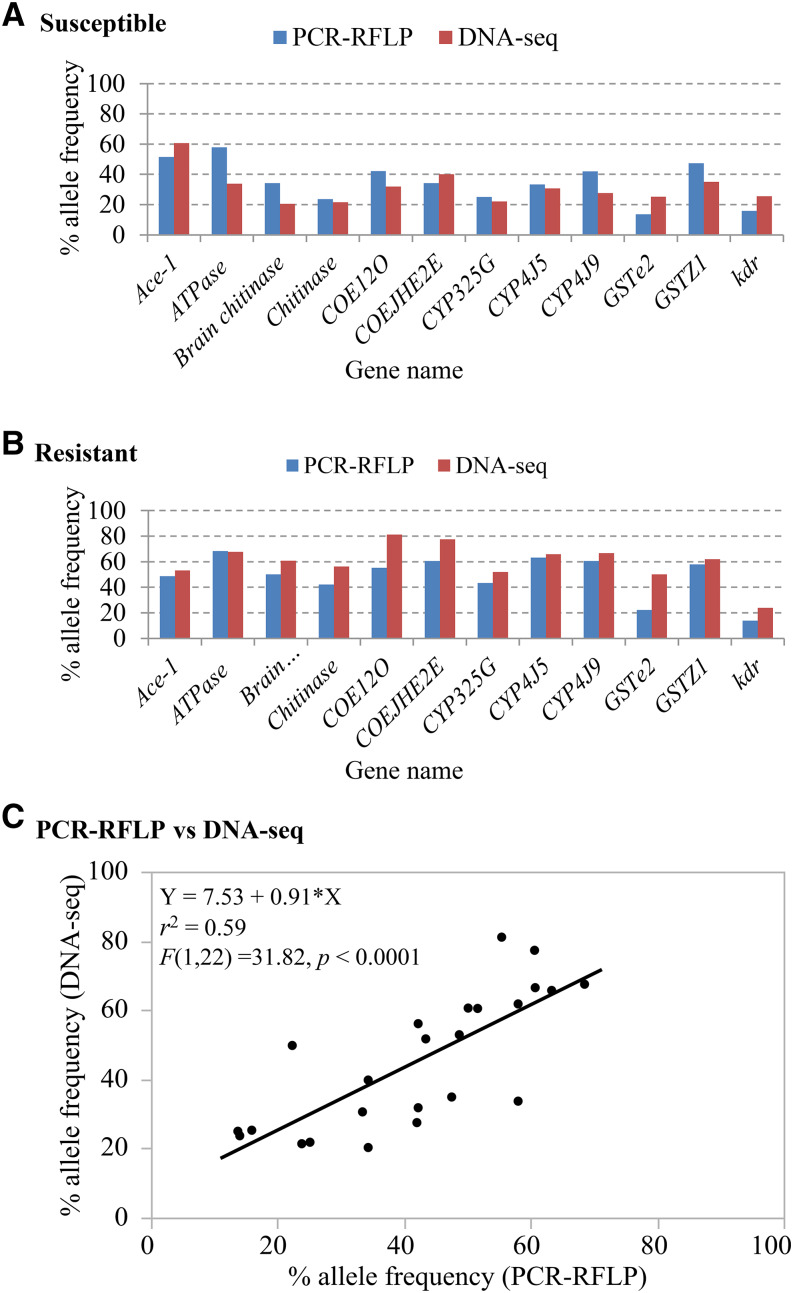
To examine the difference between pooled DNA sequencing (DNA-seq) and individually genotyping (PCR-RFLP) determined allele frequencies, an additional 40 phenotyped mosquitoes (20 resistant and 20 susceptible) from the same mosquito population were genotyped individually using PCR-RFLP methods. A list of designed primers, restriction enzymes used for PCR-RFLPs genotyping, and digestion size are shown in Table S1. Similar patterns of allele frequencies and significant correlations were observed between pooled DNA-seq and individually PCR-RFLP genotyping (Figure 5A and 5B), with a simple linear regression coefficient of r2 = 0.59, F (1, 22) = 31.82, and P < 0.001 (Figure 5C), suggesting that the allele frequencies were consistent between pooled DNA-seq and individual PCR-RFLP genotyping.
Figure 5.
Validation of SNV allele frequencies determined by pooled DNA-seq in pyrethroid susceptible and resistant Anopheles sinensis mosquitoes. A: comparison of allele frequencies determined by pooled DNA-seq and individually PCR-RFLP genotyping in susceptible mosquitoes; B: comparison of allele frequencies determined by pooled DNA-seq and individually PCR-RFLP genotyping in resistant mosquitoes; C: relationship between pooled DNA-seq and individually PCR-RFLP genotyping determined allele frequencies. A zero-allele frequency means that all alleles are reference alleles, whereas 100% allele frequency indicates that all allele are non-reference alleles.
Copy number variation (CNV) in deltamethrin -resistant and -susceptible mosquitoes
Copy number variations (CNVs) play an important role in evolution and adaptation. CNVs may contribute to insecticide resistance and affect gene structures and expression levels. Out of the 19,708 reference genes (AsinC2) included in the analysis, 56 (0.28%) were detected in copy number variation with fold changes >1.2 at an FDR adjusted p-value < 0.001. Of these 56 genes, 33 had increased gene copy numbers (amplification) in resistant mosquitoes, whereas 23 showed loss of gene copy numbers (deletion) in resistant mosquitoes (Table S5). The two genes ASIC004384 (Histidine kinase) and ASIC017541 with increased gene copy numbers showed the highest fold change (1.34), followed by ASIC004581(1.33) and ASIC011293 (1.33) in resistant mosquitoes. The two genes ASIC020925 (Protein PRRC2A isoform X1) and ASIC009866 with decreased gene copy number had the highest fold change (1.44), followed by ASIC006698 (Glutathione s-transferase E2) (1.36) in resistant mosquitoes. The other genes with CNVs included ASIC002830 (Glutaredoxin), ASIC012426 (Glycosyl transferase family 2), ASIC013265 (virulence protein), ASIC015344 (Alcohol dehydrogenase GroES domain protein), as well as 35 genes without known functions. Increased gene copy number of GSTe2 associated with insecticide resistance has been reported in several mosquito species, including Cx. quinquefasciatus (Kothera et al. 2019), Aedes aegypti (Faucon et al. 2015), and An. gambiae (Lucas et al. 2019). Interestingly, we detected a decreased trend of CNVs (fold change = -1.36) of GSTe2 in the resistant mosquitoes, suggesting that this gene might play a different role compared to those Anopheles species from Africa (Lucas et al. 2019).
Discussion
In the study, we described the whole-genome sequencing of pooled samples and subsequent identification of genetic variants in the genome of An. sinensis, an important malaria vector in Southeast Asia. Identifying genomic variants is a crucial step for unraveling the relationship between genotypes and insecticide resistance phenotypes and can yield important insights into insecticide resistance mechanisms. Our results suggested that resistance to deltamethrin in An. sinensis was not caused by the kdr mutation as reported in other mosquito species, but the differential SNVs detected in this study could play important roles in deltamethrin resistance in this species. Additionally, the detected gene copy number variations (CNVs) here may also be responsible for deltamethrin resistance. The genetic variants identified in this study represent a significant resource for future investigations into the mechanisms of An. sinensis insecticide resistance.
The newly identified genomic variants, such as SNV (Asp84Glu) in indolepyruvate ferredoxin oxidoreductase subunit alpha gene and SNV (Ser94Thr) in L-carnitine dehydratase/alpha-methylacyl-CoA racemase gene, might have some important implications in the role of pyrethroid resistance since they showed significantly higher mutation frequencies in the resistant mosquitoes. In addition, the cytochrome P450 detoxification enzyme genes, such as CYP4J5 (ASIC012923: Thr386Lys and Glu162Asp), CYP9J4 (ASIC017824: Val299Ala), and COEJHE5E(ASIC016833: Lys495Thr) were also detected with significantly higher mutation frequencies in the resistant mosquitoes, suggesting they have a role in pyrethroid insecticide resistance. Resistance-associated SNVs in P450 genes were also detected in other vector mosquito species, including An. gambiae (Weetman et al. 2018b), An. arabiensis (Lynd et al. 2019), and An. funestus (Tchigossou et al. 2018). Overexpression of CYP9J4 gene linked to insecticide resistance were found in An. gambiae (Nkya et al. 2014) and An. arabiensis (Matowo et al. 2014). Furthermore, several genes with unknown functions showed highly differential genomic variants. Further studies are needed to illustrate their roles in insecticide resistance.
This study demonstrated that pooled DNA-seq is a cost-effective and powerful tool for analysis of genome-wide allele frequency data in deltamethrin -resistant and -susceptible An. sinensis mosquitoes. However, there are also some limitations with the techniques used in this study. A pooled DNA-seq method is prone to alignment problems due to copy number variation or problems in reference genomes (Schlötterer et al. 2014). In addition, our sample size of 20 individuals for each pool resulting sequencing depth per individual of 2.1× was relatively small. Therefore, the detection of low-frequency alleles was challenging due to the difficulty in distinguishing them from sequencing errors. The pool size coupled with the amount of sequence generated could limit the detection of rare SNVs. It was only possible to detect the SNVs with 10% or higher frequency in this study. Furthermore, the technical errors in pipetting or DNA quantification may result in imbalanced pools affecting the results (Sham et al. 2002).
In summary, this was the first study describing genome variation in An. sinensis mosquitoes and comparing mutations between insecticide -resistant and -susceptible An. sinensis populations. We identified over 30 thousand non-synonymous variants with nearly one thousand differentiating SNVs as well as 56 CNVs between deltamethrin- resistant and -susceptible populations. Of these, 37 genes with function annotations belonged to the classes of immunity, detoxification, cuticular protein, and odorant protein. The genomic variations and copy number variations described here provided a useful resource for future studies of insecticide resistance mechanisms.
Acknowledgments
We thank Ying Wang, Jianhua Duan, Jiangyan Li, Ning Zhou, Tielong Xu, and Fengyang Fu for assistance with mosquito collection. We thank Erica Zhong and Seungyeon Chelsea Han for helping with DNA extraction and PCR-RFLP genotyping. We also thank the Associate Editor, Stuart Macdonald, and the anonymous reviewers for their constructive comments that improved this manuscript, and Elizabeth Hemming-Schroeder for her editorial assistance. This work is supported by grants from the National Institutes of Health of the US (U19 AI089672 and U19 AI129326), Anhui University Excellent Young Scholars Program (gxyq2017033), and the Nature Science Key Program of Colleges and Universities of Anhui Province of China (KJ2019A0320 and KJ2019A0325).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.10315898.
Communicating editor: S. Macdonald
Literature Cited
- Afrane Y. A., Zhou G., Lawson B. W., Githeko A. K., and Yan G., 2007. Life-table analysis of Anopheles arabiensis in western Kenya highlands: effects of land covers on larval and adult survivorship. Am. J. Trop. Med. Hyg. 77: 660–666. 10.4269/ajtmh.2007.77.660 [DOI] [PubMed] [Google Scholar]
- Alout H., Djegbe I., Chandre F., Djogbenou L. S., Dabire R. K. et al. , 2014. Insecticide exposure impacts vector-parasite interactions in insecticide-resistant malaria vectors. Proc. Biol. Sci. 281: 20140389 10.1098/rspb.2014.0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y., 1995. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke B. D., 2008. kdr: can a single mutation produce an entire insecticide resistance phenotype? Trans. R. Soc. Trop. Med. Hyg. 102: 524–525. 10.1016/j.trstmh.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Chandre F., Darriet F., Manguin S., Brengues C., Carnevale P. et al. , 1999. Pyrethroid cross resistance spectrum among populations of Anopheles gambiae s.s. from Cote d’Ivoire. J. Am. Mosq. Control Assoc. 15: 53–59. [PubMed] [Google Scholar]
- Chang X., Zhong D., Fang Q., Hartsel J., Zhou G. et al. , 2014. Multiple resistances and complex mechanisms of Anopheles sinensis mosquito: a major obstacle to mosquito-borne diseases control and elimination in China. PLoS Negl. Trop. Dis. 8: e2889 10.1371/journal.pntd.0002889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Wilkening S., Drechsel M., and Hemminki K., 2009. SNP_tools: A compact tool package for analysis and conversion of genotype data for MS-Excel. BMC Res. Notes 2: 214 10.1186/1756-0500-2-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M., and Koekemoer L. L., 2013. Molecular systematics and insecticide resistance in the major African malaria vector Anopheles funestus. Annu. Rev. Entomol. 58: 393–412. 10.1146/annurev-ento-120811-153628 [DOI] [PubMed] [Google Scholar]
- Cohen J., 1988. Statistical power analysis for the behavioral sciences, L. Erlbaum Associates, Hillsdale, N.J. [Google Scholar]
- David J.-P., Faucon F., Chandor-Proust A., Poupardin R., Riaz M. et al. , 2014. Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genomics 15: 174 10.1186/1471-2164-15-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabate A., Baldet T., Chandre F., Akoobeto M., Guiguemde T. R. et al. , 2002. The role of agricultural use of insecticides in resistance to pyrethroids in Anopheles gambiae s.l. in Burkina Faso. Am. J. Trop. Med. Hyg. 67: 617–622. 10.4269/ajtmh.2002.67.617 [DOI] [PubMed] [Google Scholar]
- Djogbénou L., Chandre F., Berthomieu A., Dabire R., Koffi A. et al. , 2008. Evidence of introgression of the ace-1R mutation and of the ace-1 duplication in West African Anopheles gambiae s. s. PLoS One 3: e2172 10.1371/journal.pone.0002172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., 2010. The Mosquito Fauna of Yunnan (Volumes one), Yunnan Publishing Group Corporation, Yunnan Science & Technology Press, Kunming. [Google Scholar]
- Donnelly M. J., Corbel V., Weetman D., Wilding C. S., Williamson M. S. et al. , 2009. Does kdr genotype predict insecticide-resistance phenotype in mosquitoes? Trends Parasitol. 25: 213–219. 10.1016/j.pt.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Faucon F., Dusfour I., Gaude T., Navratil V., Boyer F. et al. , 2015. Identifying genomic changes associated with insecticide resistance in the dengue mosquito Aedes aegypti by deep targeted sequencing. Genome Res. 25: 1347–1359. 10.1101/gr.189225.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanguenon V., Agossa F., Badirou K., Govoetchan R., Anagonou R. et al. , 2015. Malaria vectors resistance to insecticides in Benin: current trends and mechanisms involved. Parasit. Vectors 8: 223 10.1186/s13071-015-0833-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway J., Hawkes N. J., McCarroll L., and Ranson H., 2004. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem. Mol. Biol. 34: 653–665. 10.1016/j.ibmb.2004.03.018 [DOI] [PubMed] [Google Scholar]
- Joshi D., Park M. H., Saeung A., Choochote W., and Min G. S., 2010. Multiplex assay to identify Korean vectors of malaria. Mol. Ecol. Resour. 10: 748–750. 10.1111/j.1755-0998.2010.02835.x [DOI] [PubMed] [Google Scholar]
- Kaneko H., 2011. Pyrethroids: mammalian metabolism and toxicity. J. Agric. Food Chem. 59: 2786–2791. 10.1021/jf102567z [DOI] [PubMed] [Google Scholar]
- Kothera L., Phan J., Ghallab E., Delorey M., Clark R. et al. , 2019. Using targeted next-generation sequencing to characterize genetic differences associated with insecticide resistance in Culex quinquefasciatus populations from the southern U.S. PLoS One 14: e0218397 10.1371/journal.pone.0218397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché A. D., Harris R. B., Maliska M. E., and Linkem C. W., 2013. Comparative species divergence across eight triplets of spiny lizards (Sceloporus) using genomic sequence data. Genome Biol. Evol. 5: 2410–2419. 10.1093/gbe/evt186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas E. R., Miles A., Harding N. J., Clarkson C. S., Lawniczak M. K. N. et al. , 2019. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res. 29: 1250–1261. 10.1101/gr.245795.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd A., Gonahasa S., Staedke S. G., Oruni A., Maiteki-Sebuguzi C. et al. , 2019. LLIN Evaluation in Uganda Project (LLINEUP): a cross-sectional survey of species diversity and insecticide resistance in 48 districts of Uganda. Parasit. Vectors 12: 94 10.1186/s13071-019-3353-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd A., Oruni A., Van’t Hof A. E., Morgan J. C., Naego L. B. et al. , 2018. Insecticide resistance in Anopheles gambiae from the northern Democratic Republic of Congo, with extreme knockdown resistance (kdr) mutation frequencies revealed by a new diagnostic assay. Malar. J. 17: 412 10.1186/s12936-018-2561-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Torres D., Chandre F., Williamson M. S., Darriet F., Berge J. B. et al. , 1998. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol. Biol. 7: 179–184. 10.1046/j.1365-2583.1998.72062.x [DOI] [PubMed] [Google Scholar]
- Matowo J., Jones C. M., Kabula B., Ranson H., Steen K. et al. , 2014. Genetic basis of pyrethroid resistance in a population of Anopheles arabiensis, the primary malaria vector in Lower Moshi, north-eastern Tanzania. Parasit. Vectors 7: 274 10.1186/1756-3305-7-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. J., Dudchenko O., Kingan S. B., Koren S., Antoshechkin I. et al. , 2018. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563: 501–507. 10.1038/s41586-018-0692-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. N., Rigden D. J., Dowd A. J., Lu F., Wilding C. S. et al. , 2014. Metabolic and target-site mechanisms combine to confer strong DDT resistance in Anopheles gambiae. PLoS One 9: e92662 10.1371/journal.pone.0092662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkya T. E., Akhouayri I., Poupardin R., Batengana B., Mosha F. et al. , 2014. Insecticide resistance mechanisms associated with different environments in the malaria vector Anopheles gambiae: a case study in Tanzania. Malar. J. 13: 28 10.1186/1475-2875-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorie P. N., Ademowo O. G., Irving H., Kelly-Hope L. A., and Wondji C. S., 2015. Insecticide susceptibility of Anopheles coluzzii and Anopheles gambiae mosquitoes in Ibadan, Southwest Nigeria. Med. Vet. Entomol. 29: 44–50. 10.1111/mve.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phasomkusolsil S., Lerdthusnee K., Khuntirat B., Kongtak W., Pantuwatana K. et al. , 2011. Effect of temperature on laboratory reared Anopheles dirus Peyton and Harrison and Anopheles sawadwongporni Rattanarithikul and Green. Southeast Asian J. Trop. Med. Public Health 42: 63–70. [PubMed] [Google Scholar]
- Platt N., Kwiatkowska R. M., Irving H., Diabaté A., Dabire R. et al. , 2015. Target-site resistance mutations (kdr and RDL), but not metabolic resistance, negatively impact male mating competiveness in the malaria vector Anopheles gambiae. Heredity 115: 243–252. 10.1038/hdy.2015.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson H., Jensen B., Vulule J. M., Wang X., Hemingway J. et al. , 2000. Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Mol. Biol. 9: 491–497. 10.1046/j.1365-2583.2000.00209.x [DOI] [PubMed] [Google Scholar]
- Reimer L., Fondjo E., Patchoke S., Diallo B., Lee Y. et al. , 2008. Relationship between kdr mutation and resistance to pyrethroid and DDT insecticides in natural populations of Anopheles gambiae. J. Med. Entomol. 45: 260–266. 10.1093/jmedent/45.2.260 [DOI] [PubMed] [Google Scholar]
- Riveron J. M., Yunta C., Ibrahim S. S., Djouaka R., Irving H. et al. , 2014. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 15: R27 10.1186/gb-2014-15-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C., Tobler R., Kofler R., and Nolte V., 2014. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15: 749–763. 10.1038/nrg3803 [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Begun D. J., and Hahn M. W., 2013. Detecting highly differentiated copy-number variants from pooled population sequencing. Pac. Symp. Biocomput. 18: 344–355. 10.1142/9789814447973_0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham P., Bader J. S., Craig I., O’Donovan M., and Owen M., 2002. DNA Pooling: a tool for large-scale association studies. Nat. Rev. Genet. 3: 862–871. 10.1038/nrg930 [DOI] [PubMed] [Google Scholar]
- Srivastava H., Sharma M., Dixit J., and Das A., 2010. Evolutionary insights into insecticide resistance gene families of Anopheles gambiae. Infect. Genet. Evol. 10: 620–628. 10.1016/j.meegid.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Tchigossou G., Djouaka R., Akoton R., Riveron J. M., Irving H. et al. , 2018. Molecular basis of permethrin and DDT resistance in an Anopheles funestus population from Benin. Parasit. Vectors 11: 602 10.1186/s13071-018-3115-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toé K., N’Fale S., Dabire R., Ranson H., and Jones C., 2015. The recent escalation in strength of pyrethroid resistance in Anopheles coluzzi in West Africa is linked to increased expression of multiple gene families. BMC Genomics 16: 146 10.1186/s12864-015-1342-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D., Djogbenou L. S., and Lucas E., 2018a Copy number variation (CNV) and insecticide resistance in mosquitoes: evolving knowledge or an evolving problem? Curr. Opin. Insect Sci. 27: 82–88. 10.1016/j.cois.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D., and Donnelly M. J., 2015. Evolution of insecticide resistance diagnostics in malaria vectors. Trans. R. Soc. Trop. Med. Hyg. 109: 291–293. 10.1093/trstmh/trv017 [DOI] [PubMed] [Google Scholar]
- Weetman D., Wilding C. S., Neafsey D. E., Muller P., Ochomo E. et al. , 2018b Candidate-gene based GWAS identifies reproducible DNA markers for metabolic pyrethroid resistance from standing genetic variation in East African Anopheles gambiae. Sci. Rep. 8: 2920 10.1038/s41598-018-21265-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill M., Malcolm C., Chandre F., Mogensen K., Berthomieu A. et al. , 2004. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol. Biol. 13: 1–7. 10.1111/j.1365-2583.2004.00452.x [DOI] [PubMed] [Google Scholar]
- WHO, 2012 Global plan for insecticide resistance management in malaria vectors, World Health Organization: Geneva, Switzerland. Available at http://www.who.int/malaria/publications/atoz/gpirm/en/.
- WHO, 2013 Test procedures for insecticide resistance monitoring in malaria vector mosquitoes, World Health Organization: Geneva, Switzerland. Available at http://www.africairs.net/wp-content/uploads/2012/08/Test-procedures-for-insecticide-resistance-monitoring-WHO.pdf.
- WHO, 2019 Guidelines for malaria vector control, World Health Organization, Geneva, Switzerland. Available at https://www.who.int/malaria/publications/atoz/9789241550499/en/. [PubMed]
- Ye J., Fang L., Zheng H., Zhang Y., Chen J. et al. , 2006. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34: W293–W297. 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D., Chang X., Zhou G., He Z., Fu F. et al. , 2013. Relationship between knockdown resistance, metabolic detoxification and organismal resistance to pyrethroids in Anopheles sinensis. PLoS One 8: e55475 10.1371/journal.pone.0055475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Zhong D., Cao J., Zhou H., Li J. et al. , 2014. Transcriptome profiling of pyrethroid resistant and susceptible mosquitoes in the malaria vector, Anopheles sinensis. BMC Genomics 15: 448 10.1186/1471-2164-15-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are available in the Sequence Read Archive under the accession number SRR830401 and SRR830336 for the Illumina whole genome sequencing of genomic DNA in resistant and susceptible mosquitoes in An. sinensis. Supplemental material available at figshare: https://doi.org/10.25387/g3.10315898