Figure 3.
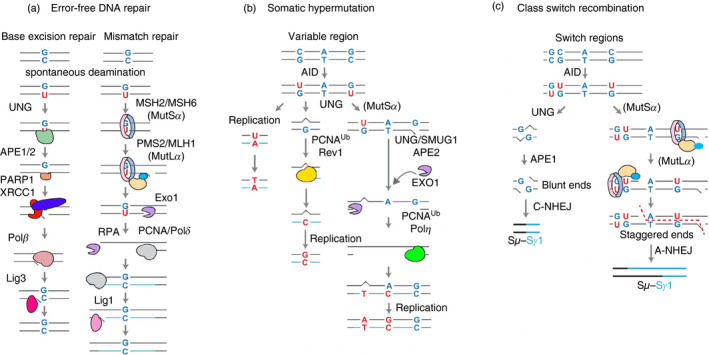
Overview of error‐free basic excision repair (BER) and mismatch repair (MMR), somatic hypermutation (SHM) and class switching recombination (CSR). (a) Error‐free repair pathways for BER (left) and MMR (right). Canonical BER is initiated by uracil‐DNA glycosylase, creating an abasic site that is recognized by APE1/2. APE1/2 nick the DNA 5′ of the abasic site. PARP1 and XRCC1 are activated and recruit proliferating cell nuclear antigen (PCNA) and polymerase β (Polβ). Polβ removes the remaining 5′ deoxyribose and insert a single nucleotide, followed by ligation with Lig3. In MMR, MutSα recognizes the U:G mispair and recruits MutLα. MutLα nicks the DNA 5′ of the mismatch via PMS2. Exo1 creates an ssDNA from the nick going past the mismatch site. PCNA subsequently recruits Polδ to fill over the gap, following ligation by Lig1. (b) During SHM, uracil can act as a template for replication leading to a C–T transition mutation. Alternatively, non‐canonical BER or MMR recruit low‐fidelity polymerases Polη through PCNA ubiquitination (PCNA‐Ub) leads to transition or transversion mutations. (c) For CSR, non‐canonical BER can lead to double strand breaks (DSB) when two uracils in opposite strands are closely spaced. MMR can process distantly spaced uracils, leading to staggered DSB. Blunt DSB are joined by canonical non‐homologous end joining, whereas staggered breaks are repaired by alternative non‐homologous end joining.
