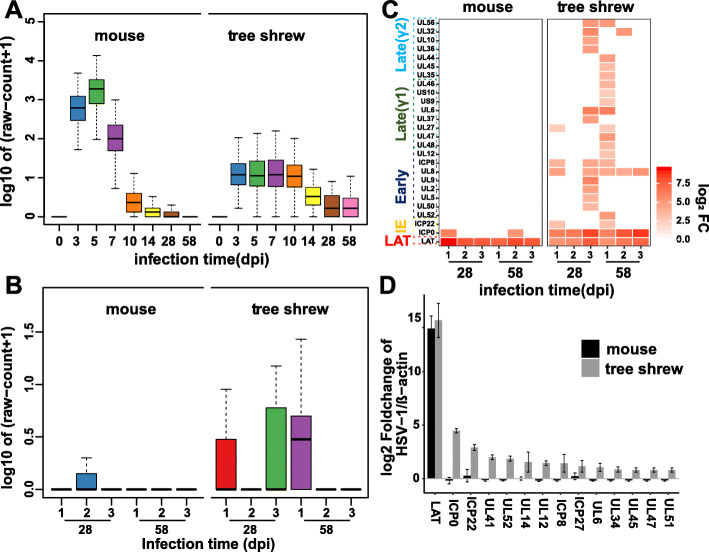
Fig. 4.
Detection of spontaneous reactivation in animal models. a Distribution of read counts for all viral genes in each time point. Viral genes with the mean raw count of three biological replicates value > 0 were selected for mapping. Each individual plot represents the value of log10 (mean + 1). b Distribution of read counts for all viral genes in each latent sample (28dpi and 58dpi), and parameter settings as described in (a). c The heat map shows viral genes that are significantly transcribed (log2 fold change > 2) in each latent sample. The graphical layout is the same as in Fig. 2a. d RT-qPCR was used to validate the viral genes in the reactivated samples. Error bars denote SD of two biological replicates. 28dpi 1# & 58dpi 2# in the mouse samples, and 28dpi 3# & 58dpi 1# in the tree shrew samples were selected for this experiment