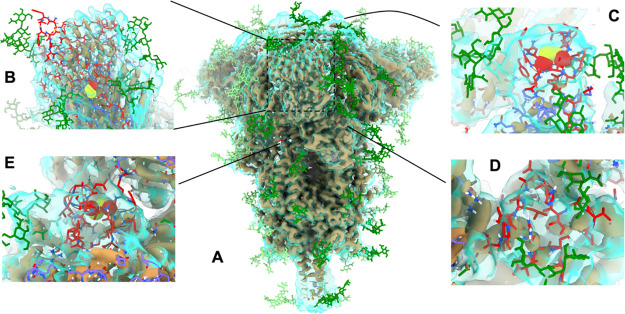
Figure 4.
Overview of key sites of extensive remodeling of 6VXX in ISOLDE. In all panels, the tan, opaque surface is the as-deposited map contoured at 9σ, while the transparent cyan surface is a smoothed map (B = 100 Å2) at 5σ. Glycans are colored in green (dark = chain A; light = chains B and C); newly-added protein carbon atoms are colored in red; pre-existing protein carbons are colored in purple (chain A) or orange (chain B). (A) Overview of highlighting sites of remodeling. (B) N-terminal domain is exceedingly poorly resolved compared to the remainder of the structure, and the distal face is essentially uninterpretable in isolation. Rebuilding was aided by reference to a crystal structure of the equivalent domain from SARS-CoV-1 (PDB: 5X4S(33)), but the result remains the lowest-confidence region of the model. Note, in particular, the addition of a disulfide bond between Cys15 and Cys136. (C) Extended loop of RBD (residues 468–489). While not visible in the high-resolution data of either cryo-EM map, the conformation seen in an RBD-ACE2 complex (PDB: 6M17(10)) was consistent with the low-resolution envelope. (D) Residues 620–641 and (E) 828–854 were associated with strong density, indicating a compact structure but too poorly resolved to support an unambiguous assignment based on the density alone. Using ISOLDE’s interactive MDFF, each loop was packed into a compact fold with reasonable geometry and a hydrophobic core, and the presence of a disulfide bond between Cys840 and Cys851 was suggested with high confidence. Illustration of S proteins and maps were generated using UCSF ChimeraX.