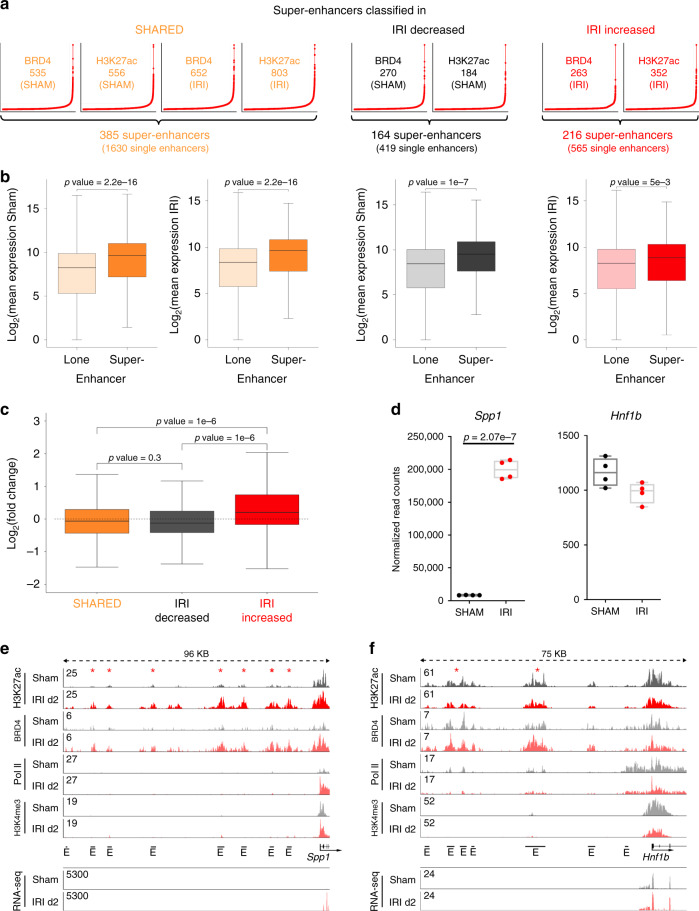
Fig. 3. Identification of super-enhancers and their annotation to genes.
a Super-enhancers were identified for all three enhancer categories using H3K27ac enhancer elements with the underlying H3K27ac and BRD4 signal. The resulting individual super-enhancers using H3K27ac or BRD4 were overlapped to identify the 385 super-enhancers based on enhancers identified in SHARED, the 164 super-enhancers based on IRI-decreased enhancers and 216 super-enhancers identified in IRI upregulated enhancers. b The box plots depict significantly higher expression for genes associated with super-enhancers. Unpaired t-tests were applied (two-sided). SHARED lone enhancer: n = 15151, SHARED super-enhancer: n = 385; IRI-decreased lone enhancer: n = 6093, IRI-decreased super-enhancer: n = 164; IRI-increased lone enhancer: n = 9209, IRI-increased super-enhancer: n = 216. Median, middle line inside each box; IQR (interquartile range), the box containing 50% of the data; whiskers, 1.5 times the IQR. c The box plot shows that genes assigned to super-enhancers from the IRI-decreased group (n = 164) have a lower gene expression level, whereas genes assigned to IRI- increased super-enhancers (n = 216) have significantly higher gene expression level compared to SHARED super-enhancers (n = 385). Median, middle line inside each box; IQR (interquartile range), the box containing 50% of the data; whiskers, 1.5 times the IQR. One-way ANOVA with Tukey post hoc test was applied. d Spp1 as a representative gene for gaining transcriptional marks is significantly upregulated at IRI d2 (based on RNA-seq data). Hnf1b is a representative gene assigned to a SHARED super-enhancer. Two-sample t-test was applied (two-sided). Individual data points and box-plot with mean ± max, min are shown. n = 4 biologically independent samples. e The induced expression of Spp1 goes along with gained enhancer elements at IRI d2. The enhancers are characterized by strongly increased H3K27ac and BRD4 coverage. The gene body shows strongly increased Pol II binding. RNA-seq track shows increased expression on exon elements after injury. f Hnf1b enhancers show mild reductions in BRD4, Pol II and H3K27ac binding. RNA-seq track shows unchanged expression on exon elements after injury.