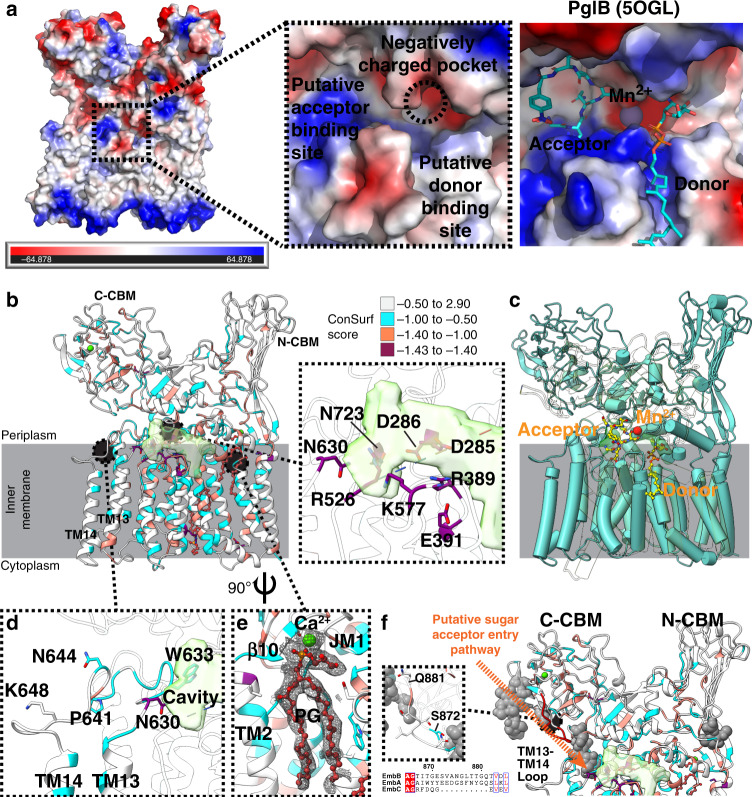
Fig. 2. Structural features of EmbB.
a Electrostatic representation of EmbB, with a zoom-in around the putative active site, labeled according to where the substrates are likely to bind, where red is more negatively charged and blue more positively charged. A comparison with the PglB active site with bound substrates is also shown, after PglB was aligned against EmbB. b Structure of EmbB, rendered in cartoon and colored based on ConSurf69 score for sequence conservation. The more negative the score, the more conserved the residue. The putative active site cavity, generated by the Voss Volume Voxelator server68 is colored in semi-transparent green. The insert shows the putative active site cavity with the strictly conserved residues labeled. c EmbB (pale blue) is superimposed on PglB (semi-transparent yellow), with the ligands and Mn2+ ion of PglB shown in yellow as sticks and a ball, respectively. d Residues that are known to maintain catalytic activity while altering substrate specificity on a loop between TM13 and TM14 are labeled and side chains are shown. e A tightly bound phosphatidylglycerol (PG shown as ball-and-stick) and calcium ion (shown as a ball) in a pocket between TM2, JM1, and β10 is shown. The density map of the lipid and the ion is displayed as mesh. f Glycan ligands for the top ten Dali server hits for both N-CBM and C-CBM were mapped onto the structure as a gray ball representation. 2WJS [10.2210/pdb2WJS/pdb] (PDB ID) was used for N-CBM, while 3PTY [10.2210/pdb3PTY/pdb] and 4GWM [10.2210/pdb4GWM/pdb] were used for the C-CBM. The missing loop in EmbC is colored in red, and the insert is a zoomed-in view. Sequence alignment of the region around the loop is appended below the insert. The putative sugar acceptor entry pathway is shown as an orange dotted line.