Figure 1. RaPID selection of K48Ubn binding cyclic peptides.
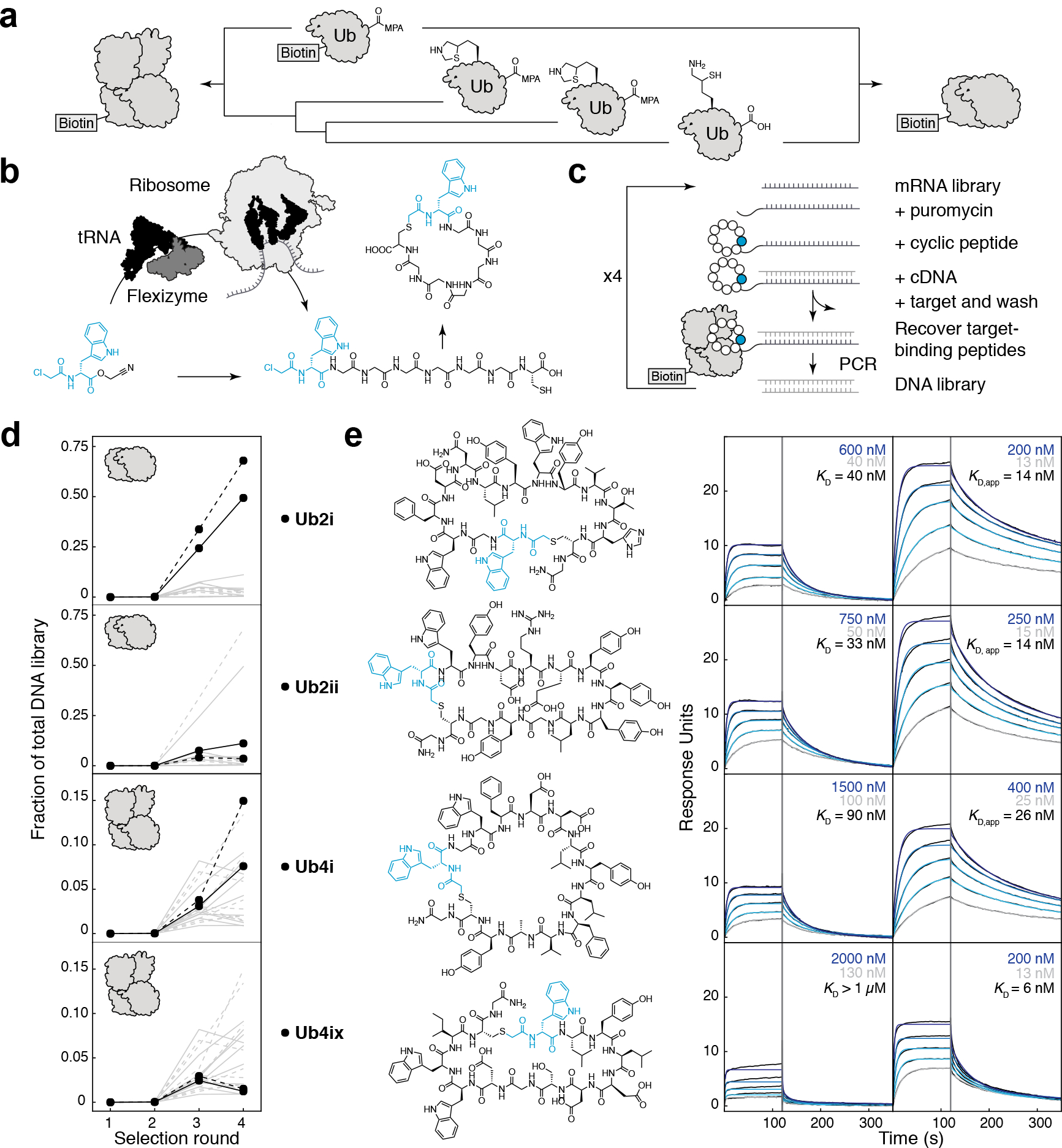
a) Chemical synthesis of biotin-K48-linked di/tetra-Ub chain. b) Flexizyme, an artificial tRNA aminoacylation ribozyme, can load nonproteinogenic amino acids onto tRNA (black), for use by the ribosome in in vitro translation. nonproteinogenic amino acids with an N-terminal ClAc group will spontaneously react with intramolecular cysteine and can be used to form macrocyclic peptides. c) Large libraries (~1013) of DNA-tagged cyclic peptides can be generated in the RaPID system, from which sequences can be isolated to tightly bind a protein target. d) Using K48Ub2 as a target, RaPID DNA libraries became enriched in certain peptide sequences (grey lines), with Ub2i and Ub2ii (black) as the dominant peptides (selection repeat shown as dashed lines). Using K48Ub4 as a target, Ub4i and Ub4ix (black) emerged amongst the enriched peptides. e) (top to bottom) Synthesized peptides Ub2i, Ub2ii, Ub4i, Ub4ix and their binding to K48Ub2 (left) and K48Ub4 (right) detected using SPR. Highest and lowest peptide SPR concentrations are shown in blue and grey respectively.
