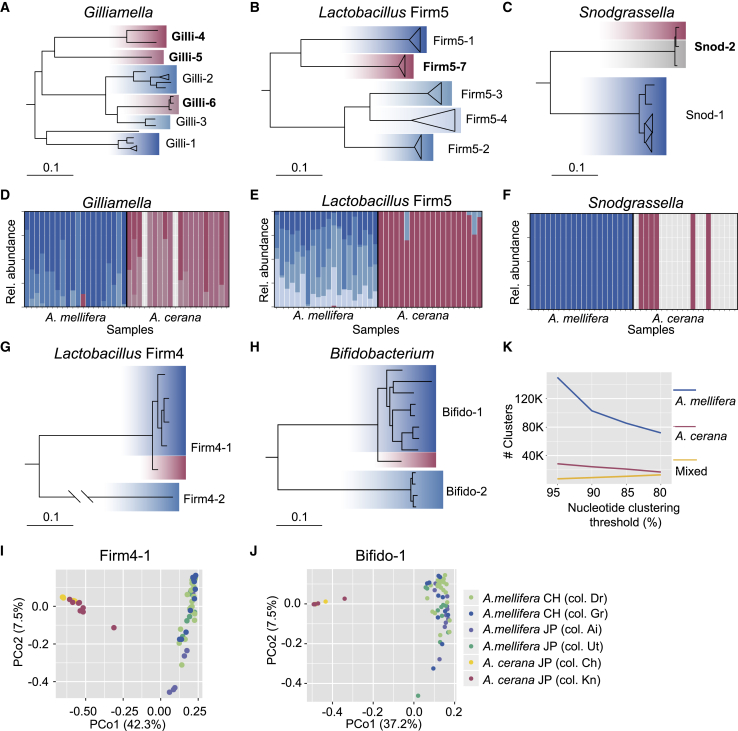
Figure 2.
The Gut Microbiota of A. mellifera and A. cerana Are Composed of Divergent SDPs and Strains
(A–C, G, and H) Core genome phylogenies of the five core phylotypes (A) Gilliamella; (B) Lactobacillus Firm5; (C) Snodgrassella; (G) Lactobacillus Firm4; and (H) Bifidobacterium; colonizing both A. mellifera and A. cerana. For (A)–(C) and (H), the trees were rooted with isolates derived from bumble bees, while for (G), the tree was rooted with the genome isolate from the Lactobacillus Firm4-2 SDP. Confirmed SDPs are indicated by the labels of the clades, with the SDPs identified in the current study highlighted with bold fonts. Genomes are highlighted with blue shades for isolates from A. mellifera and red shades for isolates from A. cerana. Gray shades indicate isolates from other honey bee species. Bars correspond to 0.1 substitutions per site.
(D–F) Barplots displaying relative abundance of the confirmed SDPs for the phylotypes (D) Gilliamella; (E) Lactobacillus Firm5; and (F) Bifidobacterium, across metagenomes from Japan.
(I and J) Principal coordinate analysis plots based on the pairwise fractions of shared SNVs (Jaccard distance) for the SDPs (I) "Firm4-1" and (J) "Bifido-1". Dots represent individual samples, color-coded by host and colony origin, as indicated by the legend. CH, Switzerland; JP, Japan.
(K) Number of host-specific and mixed sequence clusters generated from all metagenomic ORFs at different clustering thresholds. Metagenome assemblies were generated separately for each sample, using 20 million host-filtered paired-end reads per sample. Number of metagenomic samples included is as follows: n = 48 for A. mellifera and n = 20 for A. cerana.
See also Figures S2 and S3 and Data S1, S2, and S3.