Figure 5.
Possible Factors Explaining the Difference in Gut Microbiota Diversity between A. mellifera and A. cerana
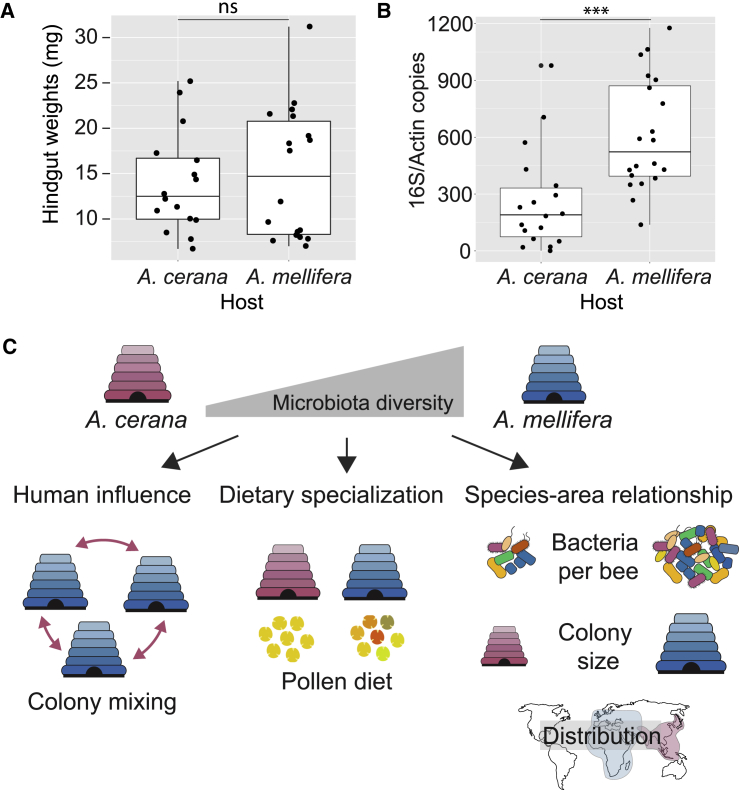
(A and B) Comparison across hosts for (A) wet weight of the hindgut (n = 18 for A. mellifera; n = 16 for A. cerana) and (B) bacterial community size (estimated with qPCR, targeting the 16S rRNA gene and normalized by copy number of the host gene actin; n = 20 for A. mellifera; n = 18 for A. cerana). For each boxplot, the center line displays the median, and the boxes correspond to the 25th and 75th percentiles; all data points are shown. Statistical significance was calculated using a Mann-Whitney U test (ns, not significant; ∗∗∗p < 0.001).
(C) Schematic illustration of three possible factors explaining differences in diversity. “Human influence”: transportation and mixing of A. mellifera colonies and genotypes around the world by beekeepers results in mixing of strains from different geographic origins and thereby increasing strain-level diversity in A. mellifera. “Dietary specialization”: A. mellifera may have a more generalist diet (here illustrated by pollen grain diversity) as compared to A. cerana and thereby be able to sustain a more diverse community. “Species-area relationship”: although previously applied to species-level diversity in animals, this concept may also apply to strain-level diversity in bacteria. For the honey bee gut microbiota, spatial differences are applicable at three levels: the size of the bacterial community within individual bees; the size of honey bee colonies; and the size of the geographic range.