Figure 2.
Validation of Mutant Screen Hits
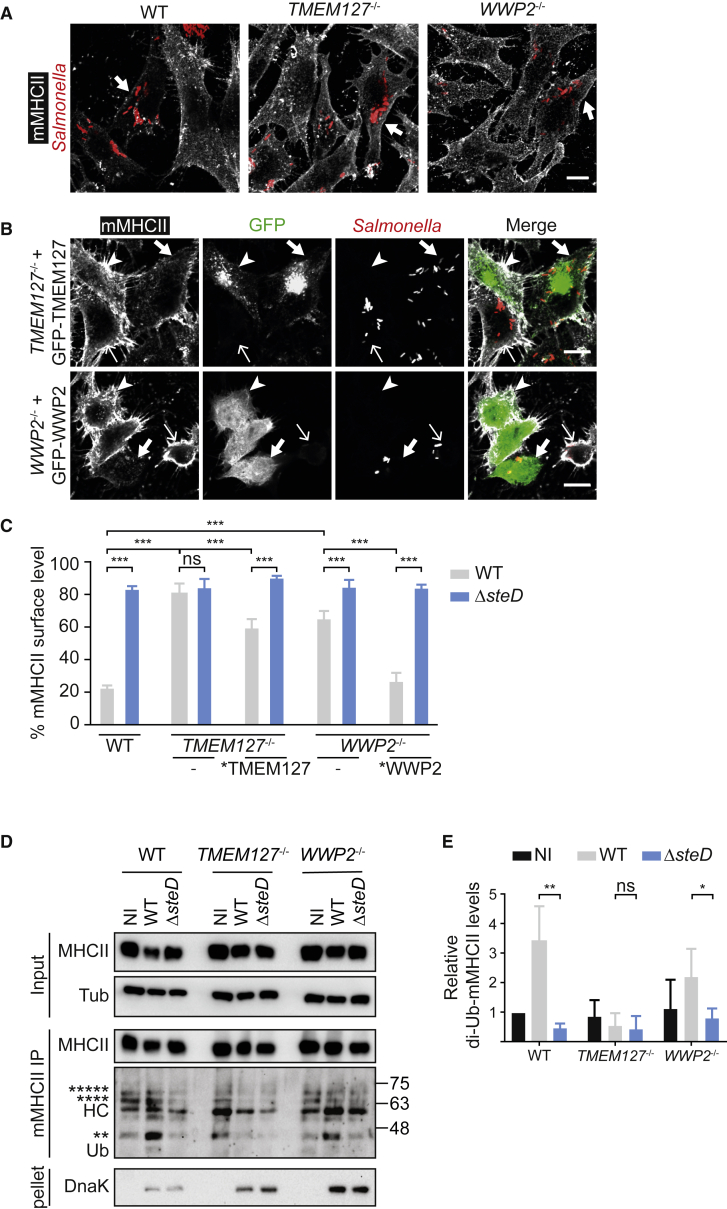
(A) TMEM127 and WWP2 are required for reduction of surface mMHCII by Salmonella. Representative confocal immunofluorescence microscopy images of wild-type (WT), TMEM127−/−, or WWP2−/− Mel Juso cells infected with mCherry-expressing WT Salmonella (red). Cells were fixed 20 h p.i. and labelled for surface mMHCII (L243 antibody, white). Arrows indicate surface mMHCII of infected cells. Scale bar, 10 μm.
(B) Mutant complementation. Representative confocal immunofluorescence microscopy images of TMEM127−/− or WWP2−/− Mel Juso cells expressing GFP-TMEM127 or GFP-WWP2, respectively (green), then infected with mCherry-expressing wild-type Salmonella (red). Cells were fixed 20 h p.i. and labelled for surface mMHCII (L243 antibody, white). Thick arrows show transfected, infected cells. Narrow arrows show non-transfected, infected cells. Arrowheads show transfected, uninfected cells. Scale bar, 10 μm.
(C) Quantification of mMHCII surface levels in mutant and complemented Mel Juso cells infected with WT or steD mutant Salmonella. Cells were analysed by flow cytometry and amounts of surface mMHCII in infected cells are expressed as a percentage of uninfected cells in the same sample. ∗TMEM127 and ∗WWP2 correspond to GFP-tagged proteins. Data are from 3 independent experiments and show means ± SD. ∗∗∗ p < 0.001, ns, not significant (two-way ANOVA followed by Tukey’s multiple comparison test).
(D) WT, TMEM127−/−, or WWP2−/− Mel Juso cells infected with WT or steD mutant Salmonella were lysed and proteins were immunoprecipitated with L243 antibody. Samples were analysed by immunoblot using anti-DRα (MHCII) anti-tubulin (Tub), anti-ubiquitin (Ub), and anti-DnaK (as a Salmonella marker) antibodies. HC – IgG heavy chain. Ubiquitin blot detects ubiquitinated mMHCII β chain (unmodified β chain –29 KDa). Bands with masses corresponding to di-, tetra-, and penta-ubiquitinated mMHCII β chain are indicated by ∗∗, ∗∗∗∗, and ∗∗∗∗∗, respectively. Protein size markers (kDa) are indicated on right.
(E) Quantification of intensity of di-ubiquitinated mMHCII signal represented in (D). Data are from 5 independent experiments and show means ± SD. ∗∗ p < 0.01, ∗ p < 0.05, ns, not significant (Student’s t test).
See also Figure S2.