Figure 6.
TMEM127- and WWP2-Dependent Ubiquitination of SteD K24
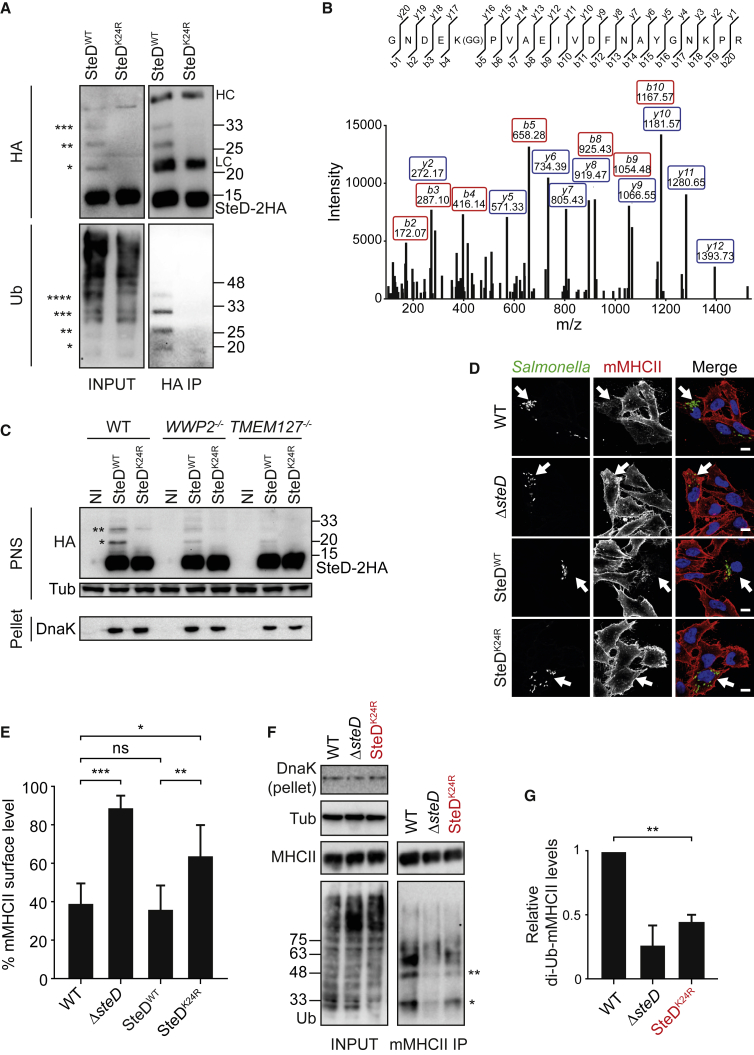
SteD variants expressed from a plasmid in steD mutant strains are indicated in black. SteDK24R expressed from the chromosome is indicated in red.
(A) SteD is ubiquitinated following translocation by intracellular Salmonella. Mel Juso cells were infected with ΔsteD Salmonella expressing HA-tagged WT SteD or K24 point mutant (SteDK24R) from a plasmid. Cells were lysed 20 h p.i. and proteins from the post-nuclear supernatant were immunoprecipitated with an HA antibody. Samples were analysed by immunoblot using anti-HA or anti-ubiquitin (Ub) antibodies. Bands with masses corresponding to mono, di-, tri-, and tetra-ubiquitinated SteD-2HA are indicated by ∗, ∗∗, ∗∗∗, and ∗∗∗∗, respectively. Protein size markers (kDa) are indicated on right. HC – IgG heavy chain. LC – IgG light chain. Representative of 3 independent experiments.
(B) MALDI MS/MS spectrum of a peptide obtained after trypsinization of GFP-SteD obtained from Mel Juso cells stably expressing GFP-SteD using GFP-trap beads. K24 is modified with a G-G branch (K[GG]) indicating ubiquitination at this position.
(C) WWP2- and TMEM127-dependent ubiquitination of SteD. WT or mutant Mel Juso cells were infected with Salmonella expressing HA-tagged WT SteD or SteDK24R from a plasmid. At 20 h p.i., proteins were separated into pellet or post-nuclear supernatant (PNS) fractions and analysed by immunoblot using anti-HA, anti-tubulin (Tub), and anti-DnaK (as a marker for Salmonella) antibodies. Bands with masses corresponding to mono and di-ubiquitinated SteD-2HA are indicated by ∗ and ∗∗, respectively. Protein size markers (kDa) are indicated on right. Representative of 3 independent experiments.
(D) SteDK24R fails to deplete surface mMHCII. Representative confocal immunofluorescence microscopy images of Mel Juso cells infected with the indicated Salmonella strains expressing GFP (green). Cells were fixed 20 h p.i. and surface mMHCII was labelled with L243 antibody (red). Nuclei were stained with DAPI (blue). Arrows indicate infected cells. Scale bar, 10 μm.
(E) Quantification of mMHCII surface levels in Mel Juso cells infected with the indicated Salmonella strains. Cells were analyzed by flow cytometry 20 h p.i., and amounts of surface mMHCII are expressed as a percentage of uninfected cells from the same sample. Data are from 3 independent experiments and show means ± SD. ∗∗∗ p < 0.001, ∗∗ p < 0.01, ∗ p < 0.05, ns, non-significant (one-way ANOVA followed by Tukey’s multiple comparison test).
(F) An SteDK24R chromosomal point mutant of Salmonella is defective for ubiquitination of mMHCII. Mel Juso cells infected with the indicated Salmonella strains were lysed 20 h p.i. and proteins were immunoprecipitated with L243 antibody. Samples were analyzed by immunoblot using anti-tubulin (Tub), and anti-DnaK (as a marker for Salmonella), anti-DRα (MHCII), and anti-ubiquitin (Ub) antibodies. Bands with masses corresponding to mono and di-ubiquitinated mMHCII β chain are indicated by ∗ and ∗∗, respectively.
(G) Quantification of intensity of di-ubiquitinated mMHCII signal relative to wild-type-infected cells, from 3 experiments represented in (F). Data are means ± SD from 3 independent experiments. ∗∗ p < 0.01 (one sample t test).
See also Figure S5.