Figure 7.
mMHCII and SteD Contain K63 Ubiquitin Linkages and Are Subject to Lysosomal Degradation
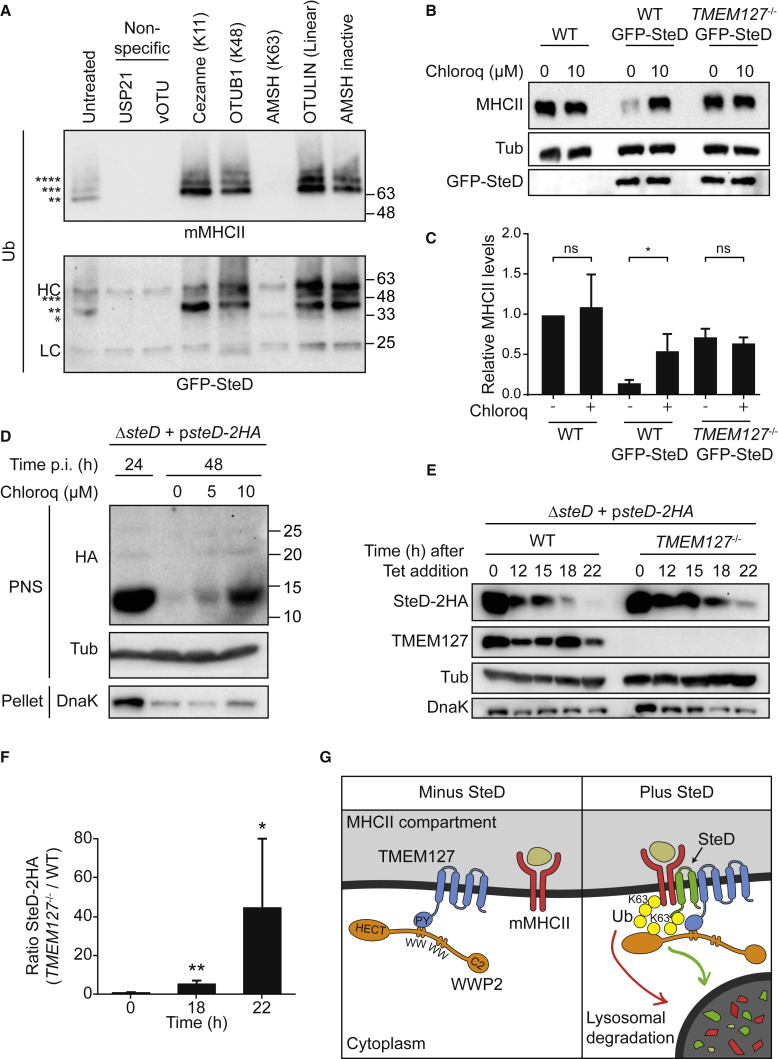
(A) UbiCRest assay of ubiquitinated mMHCII and GFP-SteD collected from Mel Juso cells stably expressing GFP-SteD. Proteins were immunoprecipitated using L243 antibody (mMHCII) or GFP-trap, incubated with the indicated DUBs and analyzed by immunoblot using a pan-ubiquitin antibody. HC – IgG heavy chain. LC – IgG light chain. Number of ∗ corresponds to predicted number of ubiquitin molecules on SteD or mMHCII. Representative of 3 independent experiments.
(B) Mel Juso cells (wild-type or TMEM127−/−), either non-transfected or expressing GFP-SteD, were mock-treated or exposed to chloroquine for 24 h. Cell lysates were analysed by immunoblot using anti-DRα (MHCII), anti-tubulin (Tub), and anti-GFP antibodies.
(C) Quantification of intensity of DRα signal from 3 experiments represented in (B), normalized to that from WT Mel Juso cells in absence of chloroquine. Data are means ± SD. ∗ p < 0.05. ns, not significant (Student’s t test).
(D) Mel JuSo cells were infected with ΔsteD Salmonella expressing SteD-2HA from a plasmid. Cells were lysed 24 h p.i. (left lane) or treated with indicated concentrations of chloroquine for an additional 24 h before lysis. Cell lysates were separated into pellet or post-nuclear supernatant (PNS) fractions and analyzed by immunoblot using anti-HA, anti-tubulin (Tub), or anti-DnaK (as a marker for Salmonella) antibodies. Representative of 3 independent experiments.
(E) WT or TMEM127−/− Mel Juso cells were infected with ΔsteD Salmonella expressing SteD-2HA from a plasmid. Bacteria were then killed with tetracycline (Tet) 20 h p.i., and the amount of SteD was analyzed at indicated time-points thereafter by immunoblot using anti-HA, anti-TMEM127, or anti-DnaK (as a marker for Salmonella) antibodies.
(F) Quantification of intensity of SteD-2HA signal at indicated times post-Tet addition. Data are means ± SD. ∗∗ p < 0.01, ∗ p < 0.05 (one sample t test).
(G) Model for mechanism of mMHCII surface depletion by SteD, TMEM127, and WWP2. TMEM127 (blue) interacts with the E3 ubiquitin ligase WWP2 (orange) via PPxY motif (PY) of TMEM127 and WW domain of WWP2. SteD (green) enables TMEM127 to interact with mMHCII (red), resulting in WWP2-dependent ubiquitination of mMHCII and SteD and their subsequent lysosomal degradation.
See also Figure S6.