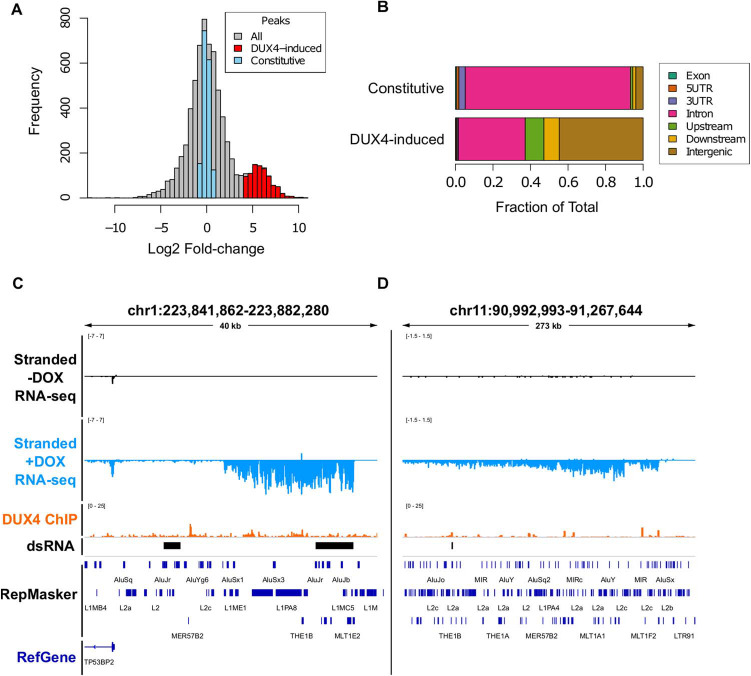
Figure 2. DUX4-induced double-stranded RNAs are enriched for non-coding intergenic RNAs. (A) Histogram of consensus ‘peaks’ from DiffBind analysis of dsRIP-seq experiment with constitutive peaks (blue, |log2 fold change| < 1.0 and FDR-adjusted P-value > 0.05),and DUX4-increased (red, log2 fold change > 4.0 and FDR-adjusted P-value ≤ 1 × 10−10). (B) Genomic feature distribution overlap of each dsRNA, defined as in Figure 2A. See Materials and Methods for feature definitions and prioritizations. (C and D) Browser screenshots in the indicated hg38 regions of normalized stranded ribominus MB135-iDUX4 RNA-seq data in two intergenic locations. These tracks represent merged read counts for the indicated duplicate datasets.