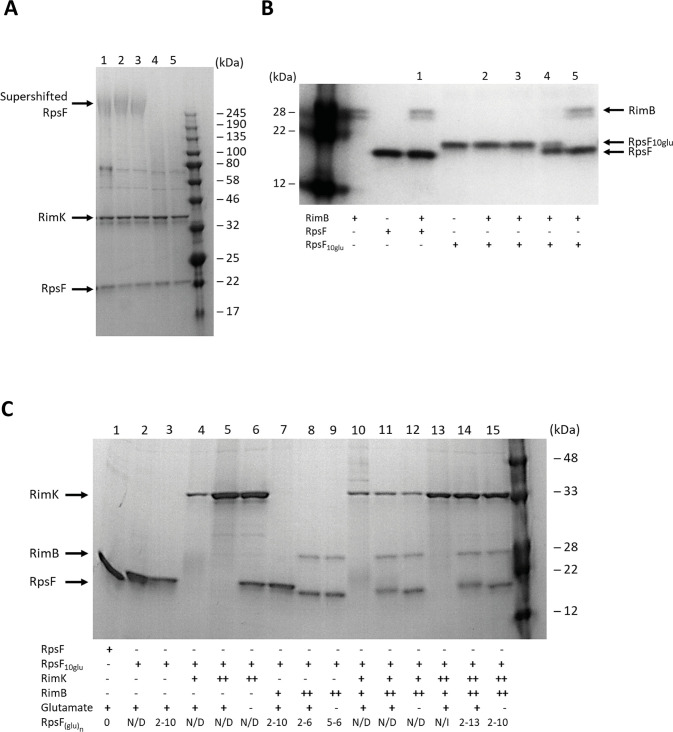
Fig 4.
(A) The presence of RimB abolishes the poly-glutamated form of RpsF. 12.5% SDS-PAGE gel. SBW25 RpsF was present at a concentration of 12.8 μM. SBW25 RimK was present at a concentration of 15 μM. Samples were incubated overnight to allow formation of the modified RpsF species shown in Lane 1. The sample was divided into four equal volumes and the following additions made: Lane 2 –buffer only; Lane 3 – 20mM ADP; Lane 4–100 nM SBW25 RimB; Lane 5–100 nM SBW25 RimB + 20 mM ADP. After 150 minutes incubation, samples were taken for SDS-PAGE analysis. The positions of the supershifted RpsF band, RimK and unmodified RpsF (Lane 1) are indicated. (B) RimB proteolyzes poly-glutamated RpsF. 12.5% SDS-PAGE gel. SBW25 RpsF and RpsF10glu were present at a concentration of 12.8 μM. RimB concentrations in the lanes labelled 1–5 were: Lane 1–6.0 μM; Lane 2–6.0 nM; Lane 3–60 nM; Lane 4–600 nM; Lane 5–6.0 μM. After 5 minutes incubation, samples were withdrawn for SDS-PAGE analysis. The positions of RimB, unmodified RpsF and RpsF10glu are indicated. (C) RimB cleaves glutamate residues from the C-terminus of RpsF. 12.5% SDS-PAGE gel. SBW25 RpsF and RpsF10glu were present at a concentration of 12.8 μM. SBW25 RimK was present at a concentration of either 3.8 μM (denoted below the gel as a single cross) or 15 μM (a double cross). SBW25 RimB was present at a concentration of either 60 nM (denoted below the gel as a single cross) or 6.0 μM (a double cross). After 60 minutes incubation, samples were withdrawn for SDS-PAGE analysis. Following SDS-PAGE, gel slices were excised from the region of the gel defined by the unmodified and partially modified RpsF bands present at the base of the gel and submitted for mass spectrometric analysis. N/D = Not Determined; N/I = Nil Identified. The positions of RimK, RimA and RpsF are indicated.