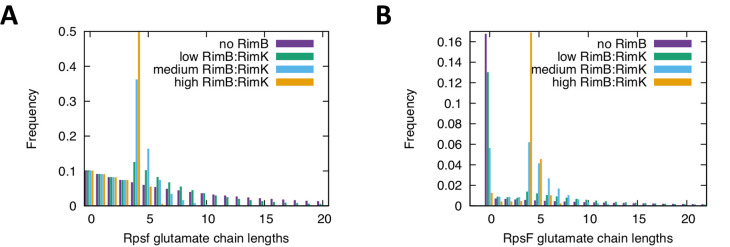
Fig 5.
(A) Changing the ratio of protease activity of RimB to RimK glutamate ligase activity changes the glutamate chain length distribution of RpsF. Stochastic simulations of glutamate addition, chain termination and chain cleavage were average over 1,000,000 simulations to obtain the expected chain length distributions as a function of a changing RimB:RimK activity ratio. As has been previously shown, with no dependency of the chain extension and termination probabilities, the chains follow an exponential distribution (purple curve, no RimB protease activity). Introducing RimB protease activity results in longer chains being reduced in length and shifting the distribution towards to minimal length at which RimB is active (four units). Increasing RimB:RimK activity further pushes the chain length distribution to peak sharply around a length of four. By changing the activity of RimK, RimA and cdG can change the ratio of protease to glutamate ligase activity in the system, with an increasing ratio sharpening the chain length distribution around four. (B) High levels of cyclic di-GMP increase the ratio of RimB protease activity to RimK glutamate ligase activity, leading to a shift towards shorter glutamate chains on RpsF and wider coverage of RpsF modification. For a limited amount of glutamate in the system and the high RpsF to RimABK ratio, the average chain lengths (Fig 5A) can lead to drastic changes in the overall modification of ribosomal units. With reduced RimB protease activity vs RimK glutamate ligase activity, a few long chains are produced on some RpsF but the majority have no chains at all (purple, no RimB protease activity). With a higher ratio of RimB protease activity vs RimK glutamate ligase activity, RimK delivers high coverage of shorter chains to more RpsF units (green to blue to orange). This protease to glutamate ligase activity is regulated by cdG and [glutamate]. Stochastic simulations were averaged from 1,000,000 runs using 500 RpsF to every RimABK.