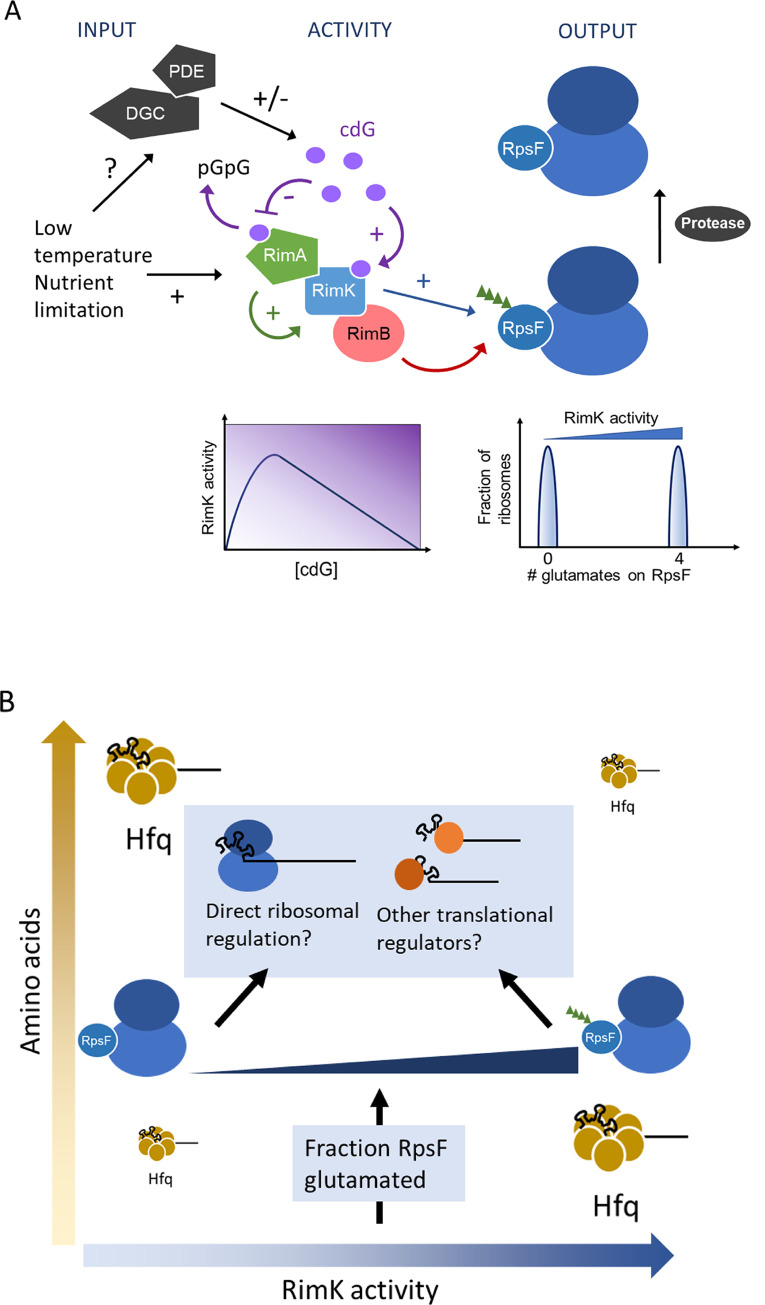
Fig 10. A model for RimABK/RpsF regulation in P. fluorescens.
Activity of RimK (blue rectangle) is stimulated by direct interaction with RimA (green pentangle) and cdG (purple circles). CdG also interacts with RimA, leading to its hydrolysis to the linear dinucleotide pGpG and an accompanying loss of RimA stimulation. The combined activity of the glutamate ligase RimK and the poly-glutamate protease RimB (red oval) adds chains of ~4 glutamate residues (green triangles) to the C-terminus of ribosomal protein RpsF (blue oval). Known inputs to the system include changes in rimABK expression, stimulated by low temperature and nutrient limitation, glutamate concentration, and cdG concentration. CdG levels are controlled in turn by as-yet-unidentified diguanylate cyclases/phosphodiesterases (grey pentangles). These inputs combine to control the proportion of glutamated ribosomes in the cell. Glutamated ribosomes are returned to a non-glutamated state by the action of an as-yet-unidentified protease (grey oval). Insets below the main diagram illustrate the impact of increasing cdG concentration on RimK activity (left, Fig 3, S6 Fig) and the predicted relationship between RimK activity and ribosomal glutamation (right, Fig 5B). (B) Model for Rim system function in high and low concentrations of amino acids. While the Rim system affects ribosome glutamation, and subsequent mRNA translation according to the model shown in 10A, the effect of the Rim system on Hfq abundance is strongly dependent on the nutrient status of the environment. This results in a regulatory output that varies in response to both RimK activity (itself controlled by cdG) and amino-acid abundance.