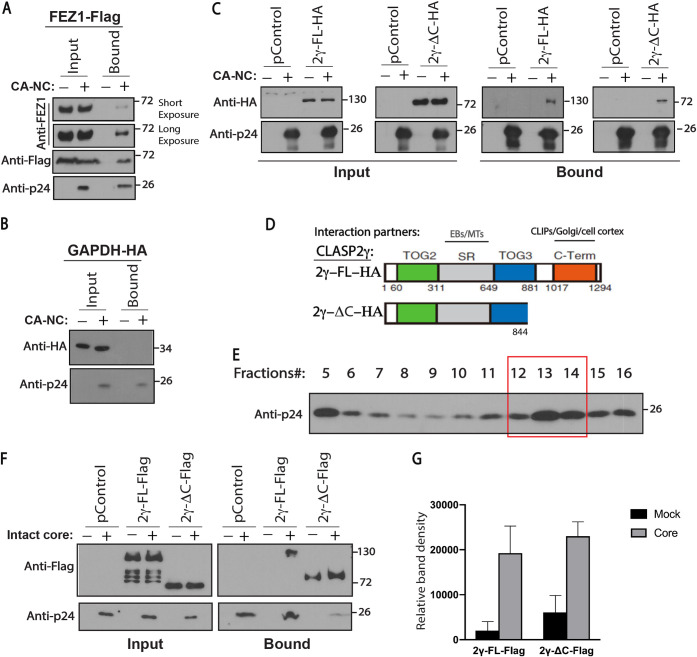
FIG 3.
CLASP2 binds to incoming HIV-1 cores. (A to C) 293T cells were transfected with plasmids expressing Flag-tagged FEZ1 (FEZ1-Flag) as the positive control (A), HA-tagged GAPDH (GAPDH-HA) (B) or empty vector (pControl) (C) as negative controls, and HA-tagged full-length CLASP2γ (2γ-FL-HA) or C-terminally truncated CLASP2γ (2γ-ΔC-HA) (C). Thirty-six hours posttransfection, cells were lysed and incubated with mock or in vitro-assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes for 1 h, followed by WB analysis of samples before (Input) or after (Bound) sedimentation through a sucrose cushion. Anti-HA and anti-p24 antibodies were used for detection of CLASP2 and HIV-1 CA, respectively. Similar results were obtained in three independent experiments. (D) Domain organization of HA-tagged full-length CLASP2γ (2γ-FL-HA) and C-terminally truncated CLASP2γ (2γ-ΔC-HA). TOG2 and -3, tumor overexpressed genes 2 and 3; SR, serine-arginine-rich sequence; C-Term, C-terminal domain. Interaction partners with SR-rich and C-terminal domains are indicated. (E and F) CLASP2 binds to intact HIV-1 cores. (E) Representative WB analysis (n = 3) showing the levels of HIV-1 p24 CA in virus fractions (only fractions 5 to 16 are shown) separated by sucrose gradient centrifugation. Fractions 5 to 7 contain free p24 not associated with the intact cores. Fractions 12 to 14 harboring intact HIV-1 cores were pooled and used in the experiment whose results are shown in panel F. (F and G) Lysates from 293T cells expressing empty vector control (pControl), Flag-tagged full-length CLASP2γ (2γ-FL-Flag), or C-terminally truncated CLASP2γ (2γ-ΔC-Flag) were incubated with fractions harboring mock (no intact core, indicated by minus signs above the gels) or intact HIV-1 cores for 2 h followed by WB analysis (F) of input and bound samples using anti-Flag and anti-p24 CA. (G) Quantification of results presented in panel F. Molecular-weight markers (in kDa) are shown to the right of WBs. Similar results were obtained in three independent experiments.